Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Behaviors of nucleosomes with mutant histone H4s
Maeshima Group / Genome Dynamics Laboratory
Kurokawa Group / Genome Evolution Laboratory
Comparative Genomics Laboratory
Behaviors of nucleosomes with mutant histone H4s in euchromatic domains of living human cells.
Adilgazy Semeigazin, Shiori Iida, Katsuhiko Minami, Sachiko Tamura, Satoru Ide, Koichi Higashi, Atsushi Toyoda A, Ken Kurokawa, Kazuhiro Maeshima *
* corresponding author
Histochemistry and Cell Biology 2024 May 14 DOI:10.1007/s00418-024-02293
DNA is a negatively charged polymer that wraps around an octamer of basic core histone proteins through electrostatic interactions, forming a nucleosome. The core histone octamer consists of eight proteins in total: two each of histones H2A, H2B, H3 and H4. Each histone has a “tail” at its terminus that lacks a specific structure and is subject to various modifications. Histone H2A has two tails, so a single nucleosome has a total of ten histone tails. Acetylation of numerous lysine residues on histone tails neutralizes their positive charges, thereby weakening their attraction to DNA and neighboring nucleosomes and causing loss of nucleosome-nucleosome interactions. A specific acetylation of histone [ histone H4 lysine-16 (H4K16Ac)] has been shown to disrupt the chromatin structure in vitro. However, it was unclear whether specific acetylation of histone tails can change the behavior of nucleosomes with that acetylation in living human cells.
A research team led by Professor Kazuhiro Maeshima of Genome Dynamics Laboratory (NIG), including a SOKENDAI graduate student Adilgazy Semeigazin (MEXT Scholar), SOKENDAI graduate students (former SOKENDAI Special Researchers, JSPS Research Fellows DC2) Shiori Iida and Katsuhiko Minami, Technical Staff Sachiko Tamura, former Assistant Professor Satoru Ide (currently Researcher at Tokyo Metropolitan Institute of Medical Science), together with Assistant Professor Koichi Higashi of Genome Evolution Laboratory (NIG), Professor Atsushi Toyoda of Comparative Genomics Laboratory (NIG), and Professor Ken Kurokawa of Genome Evolution Laboratory (NIG), analyzed the local movement of nucleosomes containing the mutation in the histone H4 tail to mimic deacetylation or acetylation. The results showed that their movement was not affected by the mutations because nucleosomes containing these mutated histones were embedded in condensed chromatin domains.
In this work, the research team ectopically expressed wild-type (wt) or mutated H4s (H4K16 point, H4K5,8,12,16 quadruple, and H4 tail deletion) fused with HaloTag in HeLa cells. Expressed wtH4-Halo, H4K16-Halo mutants, and multiple H4-Halo mutants had the euchromatin-concentrated distribution. Consistently, the genomic regions of the wtH4-Halo nucleosomes corresponded to Hi-C contact domains with active chromatin marks. Utilizing single-nucleosome imaging, the authors found that none of the H4 deacetylation or acetylation mimicked H4 mutants altered the overall local nucleosome motion. Interestingly, H4 with four lysine-to-arginine mutations displayed a substantial freely diffusing fraction in the nucleoplasm, whereas H4 with a truncated N-terminal tail was incorporated in heterochromatic regions as well as euchromatin (Figure). This study indicates the power of single-nucleosome imaging to understand individual histone/nucleosome behavior reflecting differential chemical modification states in living cells.
This year marks the centenary of the discovery of DNA staining by Robert Feulgen. This article was published in a special issue of the US journal Histochemistry and Cell Biology, “The Centennial of the Feulgen Reaction – Imaging the Genome.“
This work was supported by JSPS Fellowship, JSPS grants (24H00061, 21H02453, 20H05936, 22H05606, 23K17398, 21H02535, 23KJ0996, 23KJ0998, 22H04925 (PAGS)), a Japan Science and Technology Agency JST SPRING (JPMJSP2104), and the Takeda Science Foundation.
Video: (Left) Movement of ectopically expressed wild-type (wt) H4-Halo. Most of these were incorporated into nucleosomes and showed fluctuations. (Right) Movement of H4R-quad, a deacetylation-mimetic H4 quad mutant. They were less incorporated into the nucleosomes, with most diffusing freely.
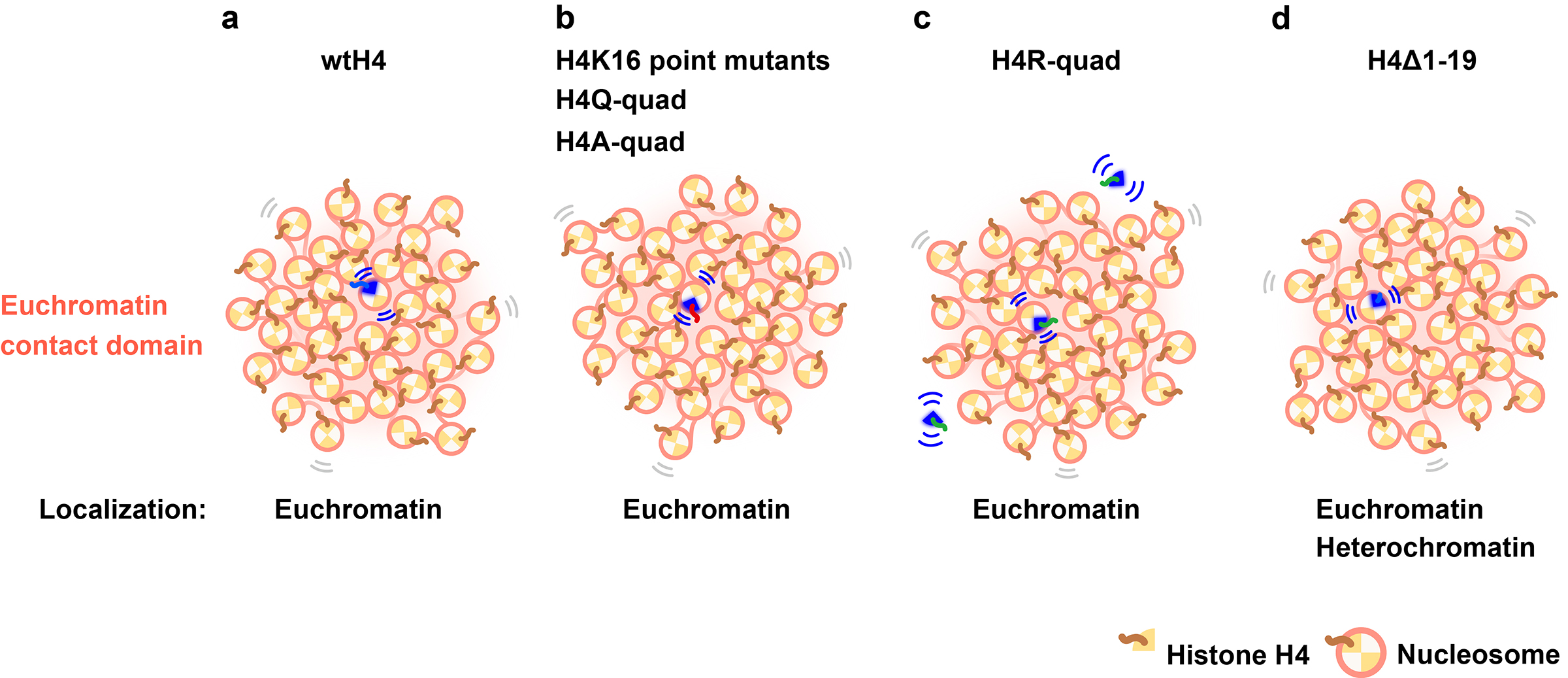
Figure: (a) Ectopically expressed wtH4-Halo was concentrated in the euchromatin contact domain. wtH4-Halo contacts adjacent nucleosomes via an H4 tail. (b) Mutations on the H4 tail (H4K16 point mutants, H4Q-quad, and H4A-quad) did not impact local nucleosome motion because the mutant nucleosomes were embedded in condensed euchromatic domains. Mutations on H4-Halo may have impeded interactions with neighboring nucleosomes, but the surrounding endogenous (wt) H4 nucleosomes were able to maintain contact with the mutant nucleosomes and thus constrain their motion. (c) H4R-quad has a nucleosome incorporation problem and has a large pool of freely diffusing H4R-quad (Movie). However, the incorporated fraction shows a similar motion as wtH4-Halo. (d) H4Δ1-19 can be incorporated into both euchromatin and heterochromatic regions. The overall motion of nucleosomes with H4Δ1-19 was lower than that of wtH4-Halo, which was primarily concentrated in the euchromatin.
Guideline for Additional Application for 2024 NIG-JOINT(Joint Researchi-(A)) (Application was closed)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Unveiling the mysteries of cell division in embryos with timelapse photography
Kanemaki Group / Molecular Cell Engineering Laboratory
Ran-GTP assembles a specialized spindle structure for accurate chromosome segregation in medaka early embryos
Ai Kiyomitsu, Toshiya Nishimura, Shiang Jyi Hwang, Satoshi Ansai, Masato T. Kanemaki, Minoru Tanaka & Tomomi Kiyomitsu
Nature Communications (2024) 15, 981 DOI:10.1038/s41467-024-45251
With the help of medaka fish, CRISPR and new imaging techniques, researchers have set a new standard for studying cell division at the very earliest stages of life.
The beginning of life is shrouded in mystery. While the intricate dynamics of mitosis is well-studied in the so-called somatic cells – the cells that have a specialized function, like skin and muscle cells – they remain elusive in the first cells of our bodies, the embryonic cells. Embryonic mitosis is notoriously difficult to study in vertebrates, as live functional analyses and -imaging of experimental embryos are technically limited, which makes it hard to track cells during embryogenesis.
However, researchers from the Cell Division Dynamics Unit at the Okinawa Institute of Science and Technology (OIST) have recently published a paper in Nature Communications, together with Professors Toshiya Nishimura from Hokkaido University (previously at Nagoya University), Minoru Tanaka from Nagoya University, Satoshi Ansai from Tohoku University (currently at Kyoto University), and Masato T. Kanemaki from the National Institute of Genetics. The study takes the first major steps towards answering questions about embryonic mitosis, thanks to a combination of novel imaging techniques, CRISPR/Cas9 genome editing technology, a modern protein-knockdown system, and medaka, or Japanese rice fish (Oryzias latipes). The timelapses that they have produced help answer fundamental questions about the intricate process of equally dividing chromosomes during embryonic mitosis, and simultaneously chart the next frontier of scientific exploration. As Professor Tomomi Kiyomitsu, senior author of the study, describes the timelapses: “they are beautiful, both on their own and because they lay a new foundation for elucidating embryonic mitosis.”
Central to the mystery of embryonic mitosis is the crucial step when the chromosomes, which contain all the genetic information of the cell, are aligned and segregated equally into daughter cells. A key player in this process is the mitotic spindle, which is made of microtubules – long protein fibers used for intra-cellular structure and transport – that radiates from opposite poles of the spindle and attaches to the chromosomes in the middle. The spindle captures duplicated chromosomes properly and segregates them equally into the daughter cells during division. There are many factors determining spindle formation, and one of these is the protein Ran-GTP, which plays an essential role in cell division of female reproductive cells, which lack centrosomes – cell organelles responsible for assembling microtubules – but not in small somatic cells, which do have centrosomes. However, it has long been unclear whether Ran-GTP is required for spindle assembly in vertebrate early embryos, which contain centrosomes but have unique features, like a larger cell size.
In contrast to mammalian early embryos, embryonic cells in fish are transparent and develop synchronously in a uniform, single-cell layer sheet, which makes them significantly easier to track. The medaka turned out to be particularly well-suited for the researchers, as these fish tolerate a wide range of temperatures, produce eggs daily, and have a relatively small genome. Being temperature-tolerant means that the medaka embryonic cells could survive at room temperature , making them particularly suited for long, live timelapse photography.
The fact that medaka produce eggs frequently and have a relatively small genome size makes them good candidates for CRISPR/Cas9-mediated genome editing. With this technology, the researchers have created genetically modified, or transgenic, medaka whose embryonic cells literally highlight the dynamics of certain proteins involved in mitosis.
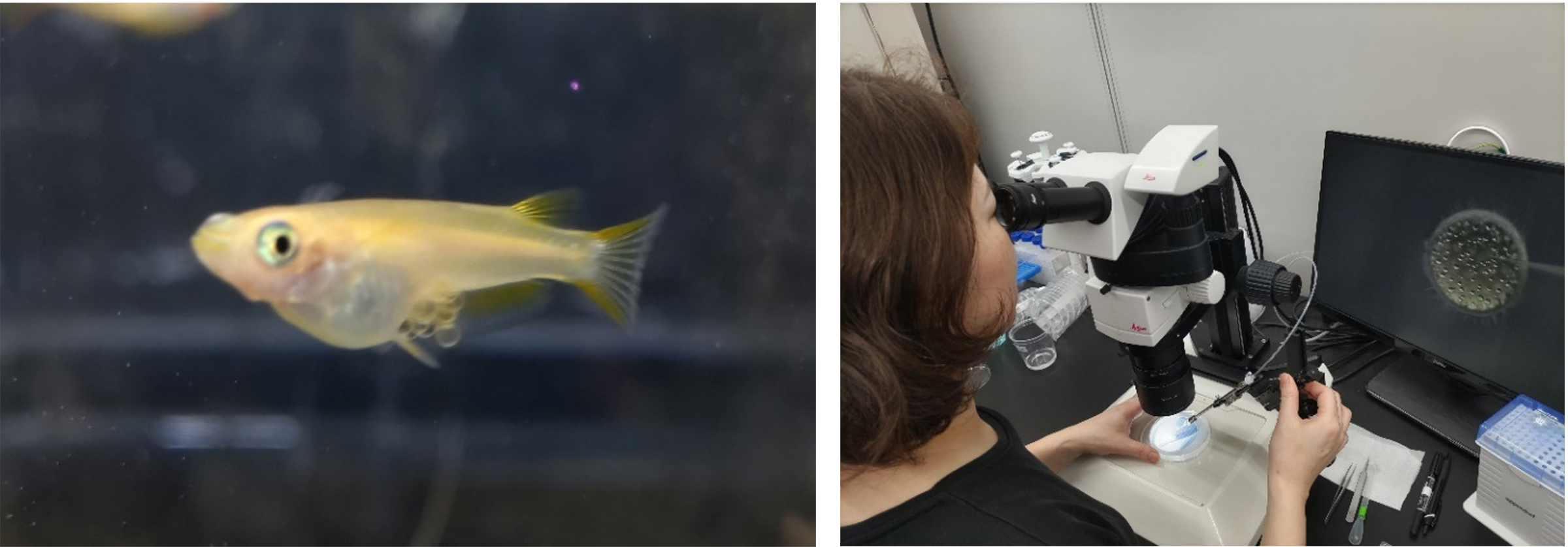
Figure: Left: Egg-carrying medaka fish, used in the study. Right: Dr. Ai Kiyomitsu from the Cell Division Dynamics Unit, first author of the paper, injecting RNA into a medaka embryo as part of the CRISPR/Cas9-mediated genome editing process. Photos: Tomomi Kiyomitsu (OIST).
In studying the timelapses of the developing mitotic spindle in live, transgenic medaka embryos, the researchers discovered that large early embryos assemble unique spindles different from somatic spindles. In addition, Ran-GTP plays a decisive role in spindle formation in early embryonic divisions, but the importance diminishes in later stage embryos. This is possibly because the spindle structure is remodeled as cells get smaller during development, though the exact reason is a subject for future research.
The researchers also discovered that the early embryonic cells do not have a dedicated spindle assembly checkpoint, which characterizes most somatic cells, and which serves to ensure that the chromosomes are properly aligned before segregation. As Professor Kiyomitsu surmises, “the checkpoint is not active, and yet the chromosome segregations are still very accurate. This could be explained by the fact that embryonic cells need to divide very quickly, but it is something that we want to study further.”
While genetically modifying the medaka fish and studying the early embryos have led to new key insights into embryonic mitosis, this is just the beginning for Professor Kiyomitsu and the team. In addition to questions related to the diminishing role of Ran-GTP in later stages and the missing spindle assembly checkpoint, he points to the satisfying symmetry of cell divisions in the timelapses: “The spindle formation is characterized by a high degree of symmetry, as the cells appear to be dividing in the sizes and defined directions, and the spindle is consistently in the center of the cells. How can the spindle orient itself so regularly across the cells, and how is it able to find the center every time?”
Moving beyond the timelapses, the team also hopes to further solidify this new foundation with additional medaka gene-lines to serve as models for research in embryonic cells, and at the same time optimize the genome editing process. Eventually, the team wants to test for generalizability of their findings by studying embryonic mitosis in other organisms, and at a later stage, they want to explore the evolution of spindle assembly and embryonic divisions, which would also contribute to a better understanding of human embryogenesis and to developing diagnosis and treatment of human infertility.
“With this paper, we have created a solid foundation,” summarizes Professor Kiyomitsu, “but we have also opened a new frontier. Embryonic mitosis is beautiful, mysterious, and challenging to study, and we hope that with our work, we can eventually get a little closer to understanding the intricate processes at the beginning of life.”

Three of the researchers involved in the study, in front of fish-tanks of the transgenic medaka fish used in the study. From left to right: Dr. Ai Kiyomitsu from OIST, Professor Satoshi Ansai from Kyoto University (previously Tohoku University), and Professor Tomomi Kiyomitsu from OIST. Photo: Tomomi Kiyomitsu.
Identifying a new functions of UHRF1 to maintain DNA methylation
KanemakiGroup / Molecular Cell Engineering Laboratory
Non-canonical functions of UHRF1 maintain DNA methylation homeostasis in cancer cells
Kosuke Yamaguchi, Xiaoying Chen, Brianna Rodgers, Fumihito Miura, Pavel Bashtrykov, Frédéric Bonhomme, Catalina Salinas-Luypaert, Deis Haxholli, Nicole Gutekunst, Bihter Özdemir Aygenli, Laure Ferry, Olivier Kirsh, Marthe Laisné, Andrea Scelfo, Enes Ugur, Paola B. Arimondo, Heinrich Leonhardt, Masato T. Kanemaki, Till Bartke, Daniele Fachinetti, Albert Jeltsch, Takashi Ito & Pierre-Antoine Defossez
Nature Communications (2024) 15, 2960 DOI:10.1038/s41467-024-47314-4
Replication of the genetic material DNA is essential for cell proliferation. During this process, not only DNA but also DNA methylation, which is essential epigenetic mark, is maintained from parent to daughter cells. It has been known that DNA methyltransferase 1 (DNMT1) and its activator, UHRF1, are important to maintain DNA methylation. Although it has been recognized primarily as an activator of DNMT1, UHRF1 is also implicated in cancer development, unlike DNMT1, suggesting that it may have functions through other pathways, independent of DNMT1.
To delineate the individual functions of UHRF1 and DNMT1, we utilized the auxin-inducible degron (AID)(In Japanese only) and AID2 system that can induce the total and synchronous depletion of endogenous UHRF1 or DNMT1 proteins in human cancer cells. Interestingly, UHRF1 depletion resulted in a more severe DNA methylation loss than DNMT1 removal. With the combination of whole genome DNA methylation analysis and genetic knock-out techniques, we revealed that UHRF1 regulated not only DNMT1 but also DNA methyltransferases DNMT3A/DNMT3B and demethylase TET2.
Assistant Prof. Kosuke Yamaguchi (formally a postdoctoral researcher at Paris-Cite University) led the study as the first and co-corresponding author in the Pierre-Antoine Defossez laboratory, and Prof. Masato Kanemaki was also involved in this study.
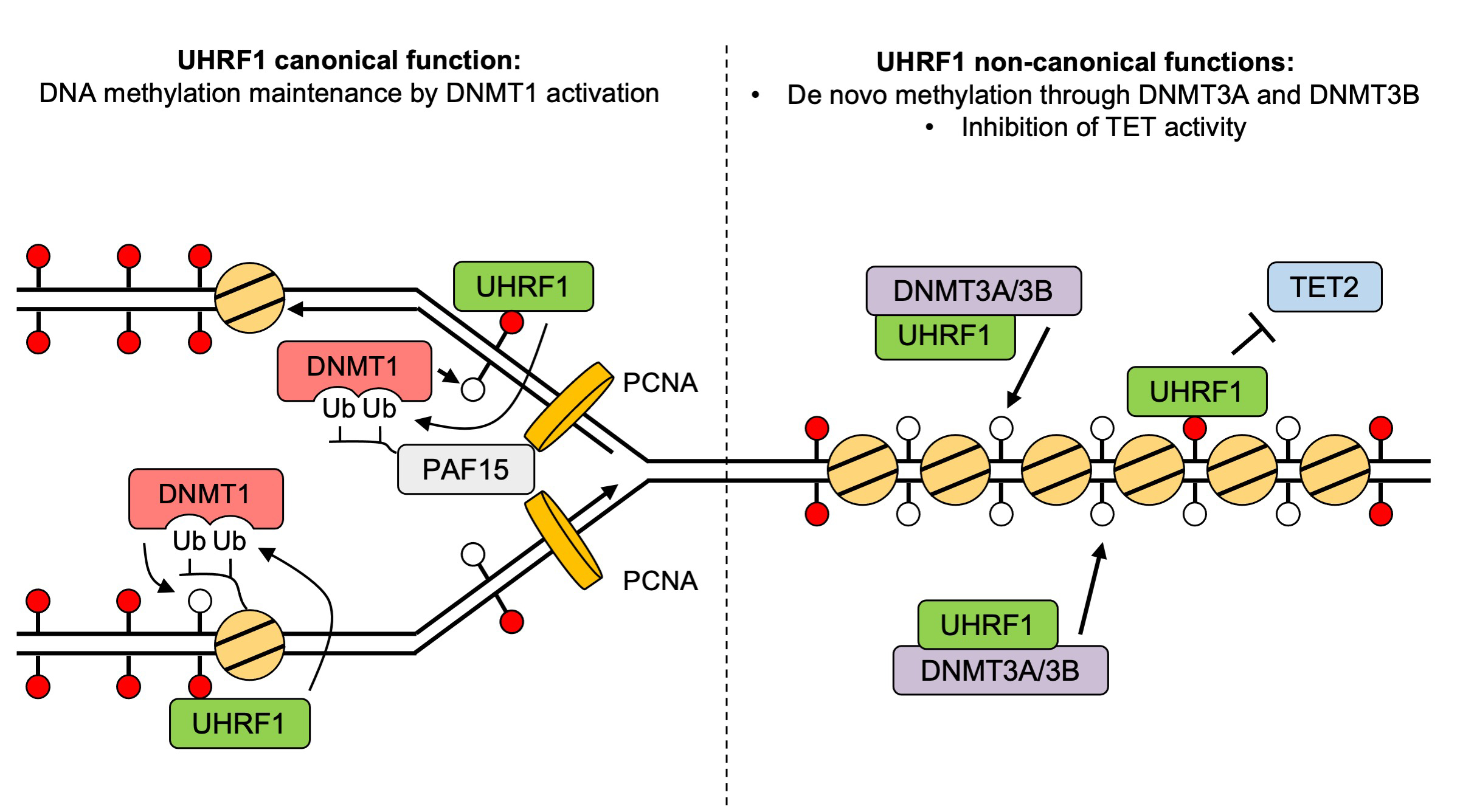
Figure: A revised and expanded model for UHRF1 functions in DNA methylation homeostasis.
Establishment of a zebrafish inbred strain, M-AB, capable of regular breeding and genetic manipulation
-Novel zebrafish bioresource from NIG-
Press release

Establishment of a zebrafish inbred strain, M-AB, capable of regular breeding and genetic manipulation
Kenichiro Sadamitsu, Fabien Velilla, Minori Shinya, Makoto Kashima, Yukiko Imai, Toshihiro Kawasaki, Kenta Watai, Miho Hosaka, Hiromi Hirata and Noriyoshi Sakai.
Scientific Reports (2024) 14, 7455 DOI:10.1038/s41598-024-57699-
![]() Press release (In Japanese only)
Press release (In Japanese only)
Inbred strains of organisms are genetically highly uniform and thus useful for life science research. We have previously reported the ongoing generation of the zebrafish IM strain from the India (IND) strain through full sib-pair mating for 16 generations. However, the IM fish laid a small number of offspring and had a short lifespan, implying the need for discreet care in breeding. Here, we report the subsequent establishment of IM strain as well as the generation of a new inbred zebrafish strain, Mishima-AB (M-AB). M-AB was derived from the *AB strain by full sib-pair mating for over 20 generations, which fulfills the general criterion for the establishment of an inbred strain. In contrast to the IM case, maintenance of the M-AB strain by sib-pair mating required almost no special handling. Genome sequencing of IM individuals from the 47th generation and M-AB individuals from the 27th generation revealed that SNP-based genomic heterogeneity across whole-genome nucleotides was 0.008% and 0.011%, respectively. These percentages were much lower than those of the parental IND (0.197%) and *AB (0.086%) strains. These results indicate that the genomes of these inbred strains were highly homogenous. We also demonstrated the successful microinjection of antisense morpholinos, CRISPR/Cas9, and foreign genes into M-AB embryos at the 1-cell stage. Overall, we report the establishment of a zebrafish inbred strain, M-AB, which is capable of regular breeding and genetic manipulation. This strain will be useful for the analysis of genetically susceptible phenotypes such as behaviors, microbiome features and drug susceptibility.
Source: Kenichiro Sadamitsu et al., Scientific Reports (2024) 14, 7455

Figure: Crossbreeding of inbred M-AB strain
A new laboratory established in the Center for Frontier Research
Dr. Kenji FUKUSHIMA joined the Center for Frontier Science as of April 1, 2024.
▶ FUKUSHIMA, Kenji : Center for Frontier Research Plant Evolution Laboratory

Associate Professor
Center for Frontier Research is an incubation center to simultaneously develop two elements: human resources and new research fields. Promising young scientists conduct research as principal investigator (tenure-track associate professor) to explore new frontiers in genetics and related areas, taking advantage of NIG’s research infrastructure and various support systems.
Three new faculty have joined NIG as of April 1, 2024
Associate Professor
Dr. Kazuhide ASAKAWA joined NIG as a Associate Professor and opened a new laboratory on April 1, 2024.
▶ ASAKAWA, Kazuhide : Neurobiology and Pathology Laboratory

ASAKAWA, Kazuhide
Associate Professor
Dr. Yasuhiro GOTO joined the Advanced Genomics Center of NIG as a Associate Professor on April 1, 2024.
▶ GOTO, Yasuhiro : Advanced Genomics Center Sequencing Division

GOTO, Yasuhiro
Associate Professor
Assistant Professor
New Assistant Professor joins NIG as of April 1, 2024.
▶ YAMAGUCHI, Kosuke : Kanemaki Group, Molecular Cell Engineering Laboratory
Second record of Macrobrachium ustulatum from Japan
Kitano Group / Ecological Genetics Laboratory
First record of Macrobrachium ustulatum (Crustacea: Decapoda: Palaemonidae) from Honshu, Japan
Yusaku Minagawa, Yusuke Fuke
Aquatic Animals (2024) 2024, .AA2024-7 DOI:10.34394/aquaticanimals.2024.0_AA2024-7
The freshwater prawn Macrobrachium ustulatum is distributed throughout the western Pacific Ocean and was first identified in Japan in 2015 on Okinawa Island, Ryukyu Islands. Here we report an adult male specimen of this species collected from the small river in Izu Peninsula, Japan. This is not only the northernmost record of M. ustulatum, but also the second record from Japan based on a single male specimen, and the first from Honshu.

Figure: Macrobrachium ustulatum obtained in this study. This is the first male specimen of this species found in Japan.
Academic publishing requires linguistically inclusive policies
Academic publishing requires linguistically inclusive policies
Henry Arenas-Castro, Violeta Berdejo-Espinola, Shawan Chowdhury, Argelia Rodríguez-Contreras, Aubrie R. M. James, Nussaïbah B. Raja, Emma M. Dunne, Sandro Bertolino, Nayara Braga Emidio, Chantelle M. Derez, Szymon M. Drobniak, Graham R. Fulton, L. Francisco Henao-Diaz, Avneet Kaur, Catherine J. S. Kim, Malgorzata Lagisz, Iliana Medina, Peter Mikula, Vikram P. Narayan, Christopher J. O’Bryan, Rachel Rui Ying Oh, Ekaterina Ovsyanikova, Katharina-Victoria Pérez-Hämmerle, Patrice Pottier, Jennifer Sarah Powers, Astrid J. Rodriguez-Acevedo, Andes Hamuraby Rozak, Pedro H. A. Sena, Nicola J. Sockhill, Anazélia M. Tedesco, Francisco Tiapa-Blanco, Jo-Szu Tsai, Jaramar Villarreal-Rosas, Susana M. Wadgymar, Masato Yamamichi, Tatsuya Amano
Proceedings of the Royal Society B: Biological Sciences (2024)291, 20232840 DOI:10.1098/rspb.2023.2840
![]() Press release (In Japanese only)
Press release (In Japanese only)
Scientific knowledge is produced in multiple languages but is predominantly published in English. This practice creates a language barrier to generate and transfer scientific knowledge between communities with diverse linguistic backgrounds, hindering the ability of scholars and communities to address global challenges and achieve diversity and equity in science, technology, engineering and mathematics (STEM). To overcome those barriers, publishers and journals should provide a fair system that supports non-native English speakers and disseminates knowledge across the globe. We surveyed policies of 736 journals in biological sciences to assess their linguistic inclusivity, identify predictors of inclusivity, and propose actions to overcome language barriers in academic publishing. Our assessment revealed a grim landscape where most journals were making minimal efforts to overcome language barriers. The impact factor of journals was negatively associated with adopting a number of inclusive policies whereas ownership by a scientific society tended to have a positive association. Contrary to our expectations, the proportion of both open access articles and editors based in non-English speaking countries did not have a major positive association with the adoption of linguistically inclusive policies. We proposed a set of actions to overcome language barriers in academic publishing, including the renegotiation of power dynamics between publishers and editorial boards.
Source: Henry Arenas-Castro et al., Proceedings of the Royal Society B: Biological Sciences (2024) vol, pp.
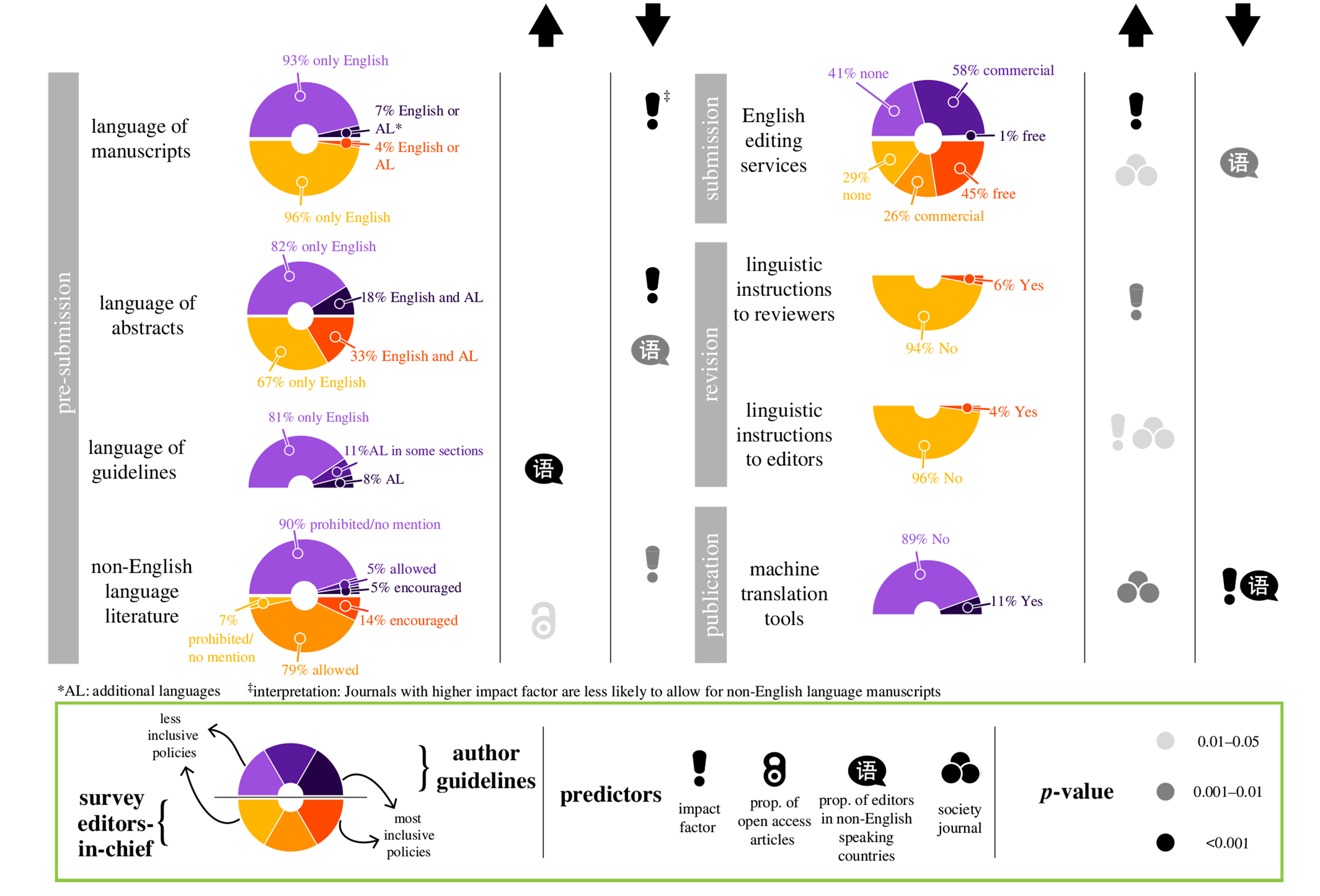
Figure: Panorama and drivers of linguistic inclusivity in academic publishing. Linguistic policies of journals as communicated in author guidelines (n = 736, the upper half of the donut) and answered in our survey by editors-in-chief (n = 262, the lower half) alongside the predictors that are associated either positively (upward arrow) or negatively (downward arrow) with the level of linguistic inclusiveness in policies. Source: Figure 1 in the article of Henry Arenas-Castro et al.
Professor Masato Kanemaki awarded the 23rd Yamazaki-Teiichi Prize
Professor Masato Kanemaki of Molecular Cell Engineering Laboratory has been awarded the 23rd Yamazaki-Teiichi Prize. This award, presented by Foundation for Promotion of Materials Science and Technology (MST), is given to individuals who have made outstanding creative achievements which lead to practical applications through the publication of technical papers, acquisition of patents, development of methods and technology. The prize is presented in four fields: “Materials,” “Semiconductors and Systems, Information, and Electronics,” “Measurement and Evaluation,” and “Bio and Medical Sciences.” In the fiscal year 2023, selection was conducted in the fields of “Measurement and Evaluation” and “Bio and Medical Sciences,” and Professor Kanemaki was selected as the recipient in the field of Bio and Medical Sciences.
https://www.mst.or.jp/Portals/0/prize/japanese/winners/newwinners.html
The prize ceremony and acceptance lecture were held on February 28, 2024, at The Japan Academy in Tokyo.



The dwarf neon rainbowfish Melanotaenia praecox, a small spiny-rayed fish with potential as a new Acanthomorpha model fish: II. Establishment of a microinjection procedure for genetic engineering
Kawakami Group / Laboratory of Molecular and Developmental Biology
The dwarf neon rainbowfish Melanotaenia praecox, a smallspiny-rayed fish with potential as a new Acanthomorphamodel fish: II. Establishment of a microinjection procedurefor genetic engineering.
Kazuhide Miyamoto, Gembu Abe, Koichi Kawakami, Koji Tamura, Satoshi Ansai
Developmental Dynamics 2024 Feb 5 DOI:10.1002/dvdy.698
Background: Rainbowfish is a clade of colorful freshwater fish. Melanotaenia praecox is a small rainbowfish species with biological characteristics that make it potentially useful as an experimental model species. We anticipate that M. praecox could become a new model used in various fields, such as ecology, evolution, and developmental biology. However, few previous studies have described experimental set-ups needed to understand the molecular and genetic mechanisms within this species. Results: We describe detailed procedures for genetic engineering in the rainbowfish M. praecox. By using these procedures, we successfully demonstrated CRISPR/Cas-mediated knockout and Tol2 transposon-mediated transgenesis in this species. Regarding the CRISPR/Cas system, we disrupted the tyrosinase gene and then showed that injected embryos lacked pigmentation over much of their body. We also demonstrated that a Tol2 construct, including a GFP gene driven by a ubiquitous promoter, was efficiently integrated into the genome of M. praecox embryos. Conclusions: The establishment of procedures for genetic engineering in M. praecox enables investigation of the genetic mechanisms behind a broad range of biological phenomena in this species. Thus, we suggest that M. praecox can be used as a new model species in various experimental biology fields.
This study was conducted as collaboration with Tamura lab at Tohoku University.
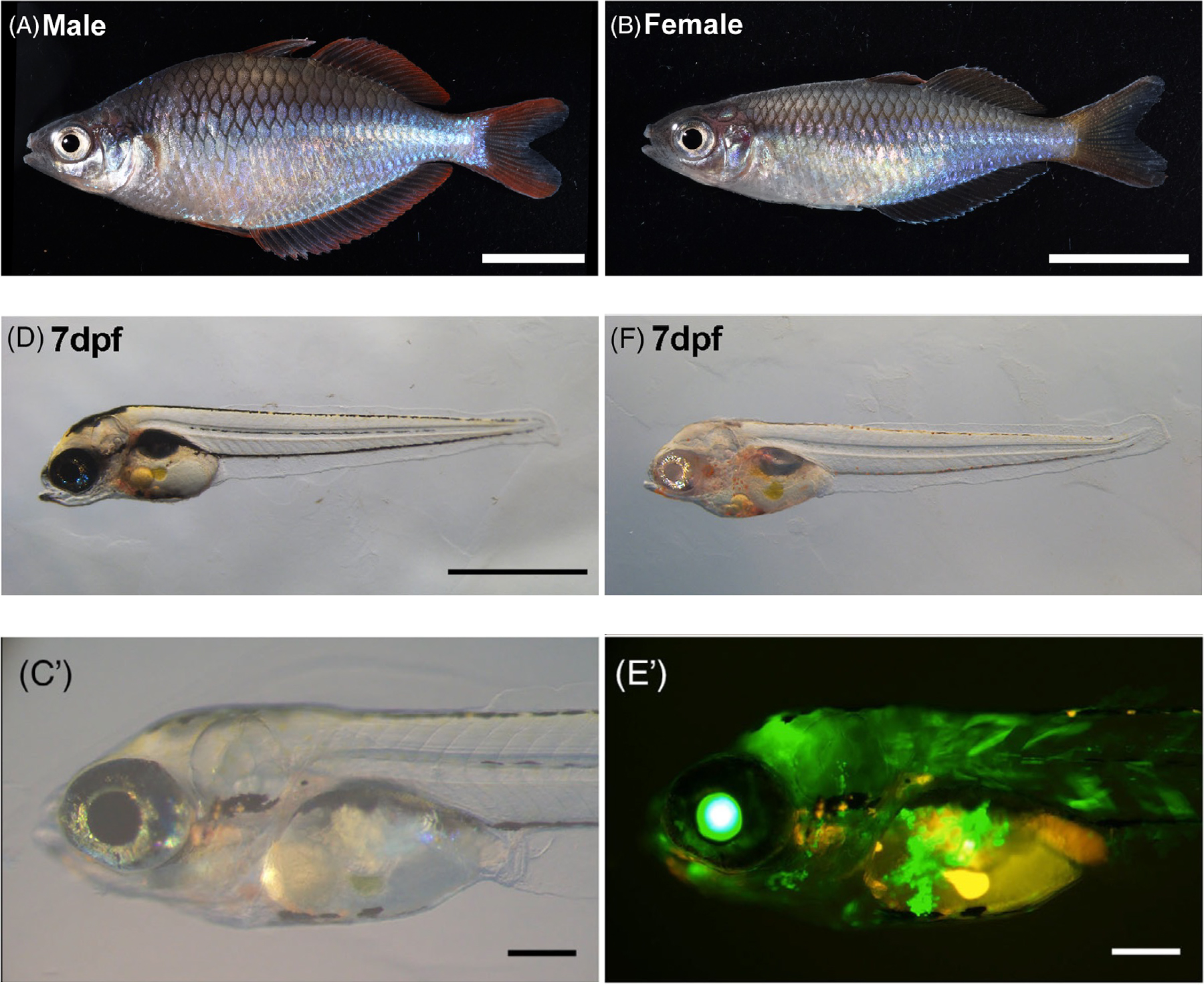
Figure: (A)(B) Male and female Melanotaenia praecox.
(D)(F) Disruption of the tyrosinase gene using CRISPR/Cas9.
(C’)(E’) Introduction of the GFP gene using Tol2.
More constrained chromatin in oncogenic HRAS-transformed cells.
Maeshima Group / Genome Dynamics Laboratory
Kurokawa Group / Genome Evolution Laboratory
Kuraku Group / Molecular Life History Laboratory
Chromatin organization and behavior in HRAS-transformed mouse fibroblasts
Aoi Otsuka*, Katsuhiko Minami*, Koichi Higashi, Akane Kawaguchi, Sachiko Tamura, Satoru Ide, Michael J. Hendzel, Ken Kurokawa, Kazuhiro Maeshima#
* These authors equally contributed to this work. #corresponding author
Chromosoma 2024 Feb 24 DOI:10.1007/s00412-024-00817-x
Genomic DNA is organized three-dimensionally in the nucleus as chromatin. Recent super-resolution imaging and Hi-C studies have shown that chromatin in living cells forms a number of condensed chromatin domains. Inside these domains, nucleosomes behave locally like a liquid, and such behavior is deeply related to various DNA functions. Cancer cells often show upregulated transcription and other activities, but how is the chromatin behavior in cancer cells different from non-cancer cells?
A research team led by Professor Kazuhiro Maeshima of Genome Dynamics Laboratory (NIG), including a SOKENDAI graduate student Aoi Otsuka (SOKENDAI Special Researcher), SOKENDAI graduate student Katsuhiko Minami (former SOKENDAI Special Researcher, JSPS Research Fellow DC2), Assistant Professor Satoru Ide, Technical Staff Sachiko Tamura, together with Assistant Professor Koichi Higashi and Professor Ken Kurokawa of Genome Evolution Laboratory (NIG), Assistant Professor Akane Kawaguchi of Molecular Life History Laboratory (NIG), and Professor Michael J. Hendzel of University of Alberta, have found chromatin in oncogenic RAS-transformed cells is more constrained than their parental cells.
In this work, the research team examined the chromatin behavior in mouse oncogenic HRAS-transformed cells (CIRAS-3 cells) and their parental cells (10T1/2 cells) with multiple approaches. First, they found that the CIRAS-3 cells have smaller nuclei. In addition, they investigated the individual nucleosome movements in living cells using super-resolution fluorescence microscopy and revealed that nucleosomes are locally more constrained in CIRAS-3 cells. Consistently, CIRAS-3 cells show increased heterochromatin, and in situ Hi-C revealed enriched interactions of the B-B compartments, which mainly consist of heterochromatin.
Genome chromatin in cancer cells encounters physical stress during migration through small spaces, such as metastasis. The smaller nuclei and the more constrained chromatin with enriched heterochromatin can gain elastic properties of chromatin, and may better benefit the cell metastasis (Figure).
This work was supported by JSPS Fellowship, JSPD grants (21H02453, 23K17398, 23K05798, 22H05606, 21H02535, 23KJ0998, 20H05936, 22H04925 (PAGS)), a Japan Science and Technology Agency JST SPRING(JPMJSP2104), The Naito Foundation, and the Takeda Science Foundation. Genome analysis was performed with support of Platform for Advanced Genome Science (22H04925 (PAGS)).
The journal “Chromosoma” is a historical chromosome journal first published in 1939, and is available from the first issue in the NIG Library.
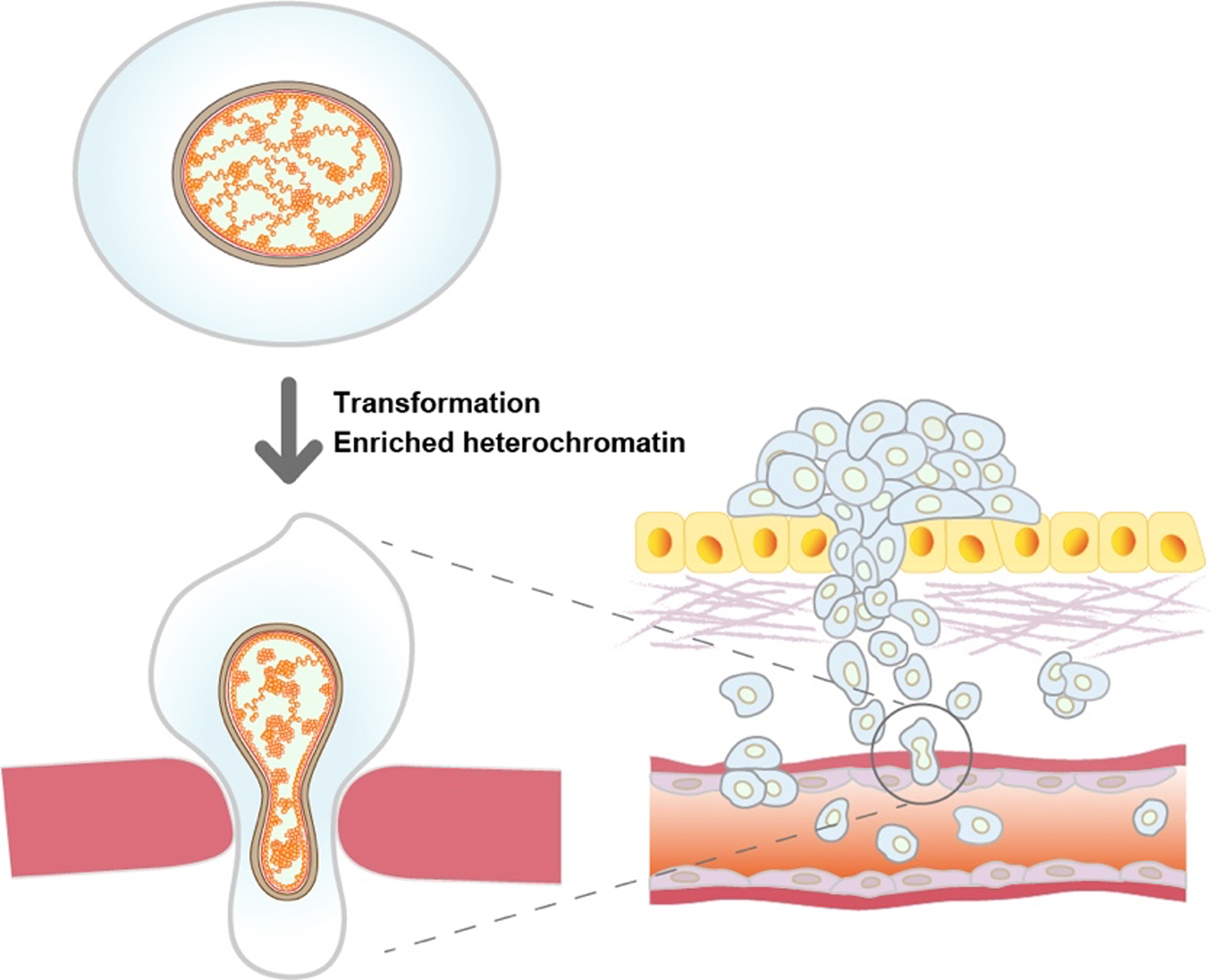
Figure: Oncogenic HRAS-transformed cells (bottom left) have more constrained chromatin with increased heterochromatin than non-transformed parental cells (top left). The more constrained chromatin may play an important role in the metastasis process.
Association of tameness and sociability but no sign of domestication syndrome in mice selectively bred for active tameness
KoideGroup / Mouse Genomics Resource Laboratory
Association of tameness and sociability but no sign of domestication syndrome in mice selectively bred for active tameness
Bharathi Venkatachalam, Bhim B. Biswa, Hiromichi Nagayama, Tsuyoshi Koide*
*責任著者
Genes, Brain and Behavior (2024) 23, e12887 DOI:10.1111/gbb.12887
Domesticated animals have been developed by selecting desirable traits following the initial unconscious selection stage, and now exhibit phenotypes desired by humans. Tameness is a common behavioural trait found in all domesticated animals. At the same time, these domesticated animals exhibit a variety of morphological, behavioural, and physiological traits that differ from their wild counterparts of their ancestral species. These traits are collectively referred to as domestication syndrome. However, whether this phenomenon exists is debatable. Previously, selective breeding has been used to enhance active tameness, a motivation to interact with humans, in wild heterogeneous stock mice derived from eight wild inbred strains. In the current study, we used tame mice to study how selective breeding for active tameness affects behavioural and morphological traits. A series of behavioural and morphological analyses on mice showed an increased preference for social stimuli and a longer duration of engagement in non-aggressive behaviour. However, no differences were observed in exploratory or anxiety-related behaviours. Similarly, selection for tameness did not affect ultrasonic vocalisations in mice, and no changes were observed in known morphological traits associated with domestication syndrome. These results suggest that there may be a link between active tameness and sociability and provide insights into the relationship between tameness and other behaviours in the context of domestication.
Source: Bharathi Venkatachalam et al., Genes, Brain and Behavior (2024) 23, e12887
Ensuring personal space inside the cell
Kimura Group / Cell Architecture Laboratory
Enucleation of the C. elegans embryo revealed dynein-dependent spacing between microtubule asters
Fujii, K., Kondo, T. & *Kimura
*corresponding author
Life Sci. Alliance (2023) 7, e202302427 DOI:10.26508/lsa.202302427
In psychology, personal space is known as the distance that people tend to maintain from others. A similar phenomenon can be found inside the cell. An organelle called centrosome tend to maintain certain distance from the other centrosomes. In this study, Dr. Ken Fujii and his colleagues investigated the behavior of the centrosomes in enucleated, C. elegans embryonic cells. Using genetics and computational approaches, the researchers proposed that the centrosomes compete with each other for motor proteins “dynein” distributed in the cytoplasm and at the cell cortex to maintain certain distance from the others and ensure their “personal space.” The study has implication in how cellular structures measure distances inside the cell.
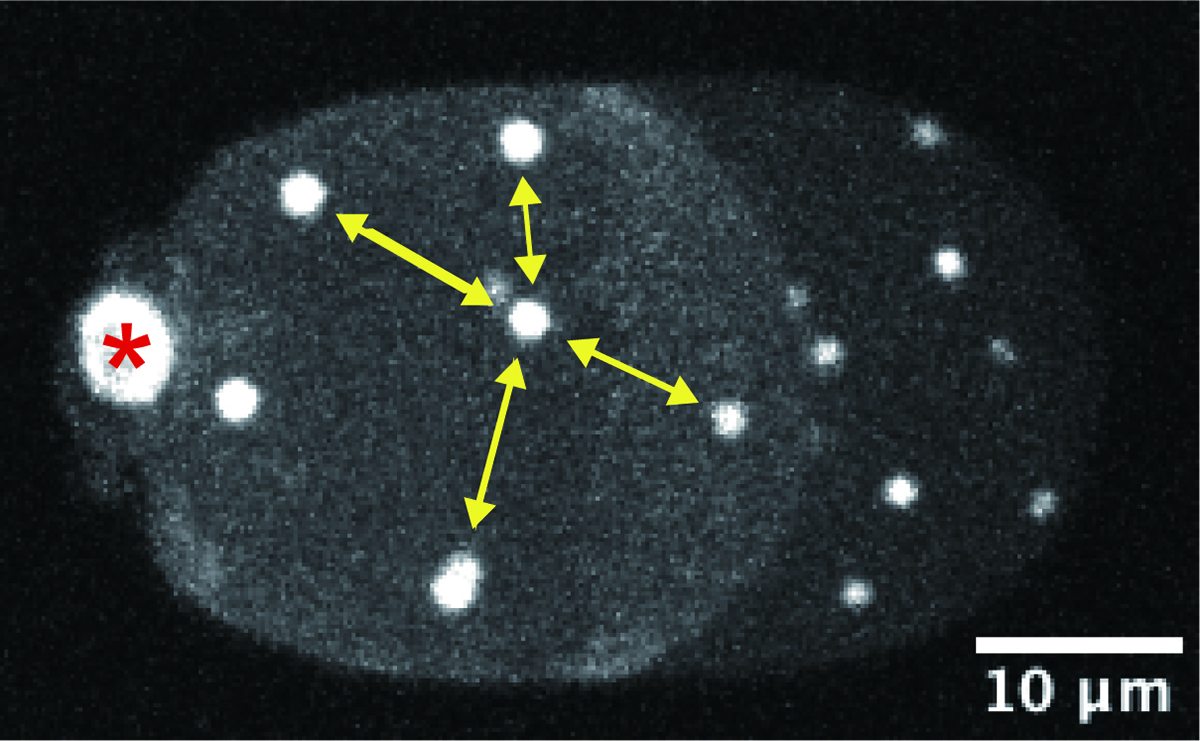
Figure: A representative microscope image of an enucleated, C. elegans embryonic cell. White signals indicate the centrosomes, which are evenly distributed inside the cell. (The image is a 2-dimensional projection of the 3-dimensional cell image. The asterisk indicates the polar body, which is not the centrosome.)
Target-selective vertebrate motor axon regeneration depends on interaction with glial cells at a peripheral nerve plexus
Kawakami Group / Laboratory of Molecular and Developmental Biology
Target-selective vertebrate motor axon regeneration depends on interaction with glial cells at a peripheral nerve plexus
Lauren J. Walker, Camilo Guevara, Koichi Kawakami, and Michael Granato
PLOS Biology (2023) 21, e3002223 DOI:10.1371/journal.pbio.3002223
A critical step for functional recovery from peripheral nerve injury is for regenerating axons to connect with their pre-injury targets. Reestablishing pre-injury target specificity is particularly challenging for limb-innervating axons as they encounter a plexus, a net-work where peripheral nerves converge, axons from different nerves intermingle, and then resort into target-specific bundles. Here, we examine this process at a plexus located at the base of the zebrafish pectoral fin, equivalent to tetrapod forelimbs. Using live cell imaging and sparse axon labeling, we find that regenerating motor axons from 3 nerves coalesce into the plexus. There, they intermingle and sort into distinct branches, and then navigate to their original muscle domains with high fidelity that restores functionality. We demonstrate that this regeneration process includes selective retraction of mistargeted axons, suggesting active correction mechanisms. Moreover, we find that Schwann cells are enriched and associate with axons at the plexus, and that Schwann cell ablation during regeneration causes profound axonal mistargeting. Our data provide the first real-time account of regenerating vertebrate motor axons navigating a nerve plexus and reveal a previously unappreciated role for Schwann cells to promote axon sorting at a plexus during regeneration.
This study was conducted as collaboration with Granato lab at University of Pennsylvania.
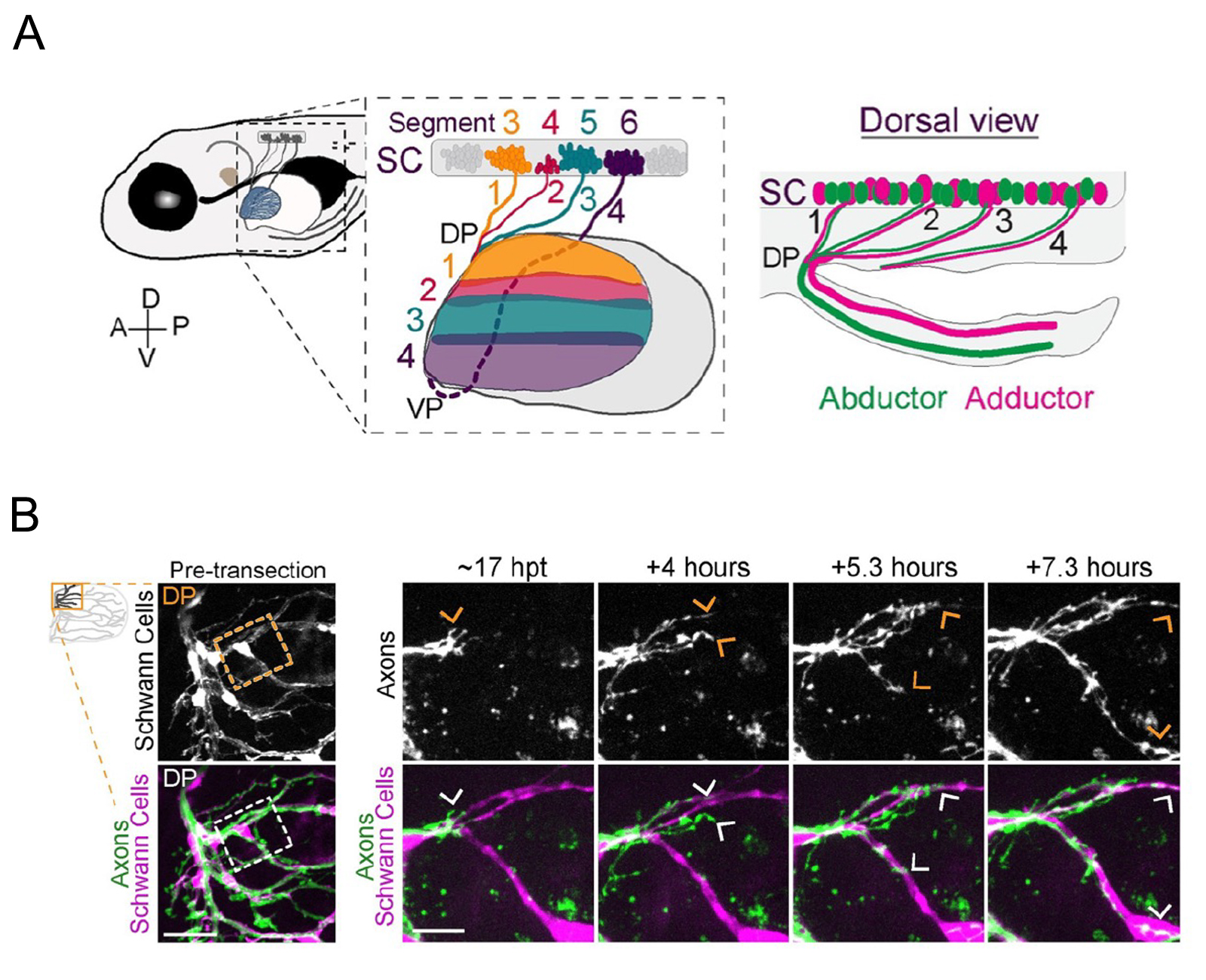
Figure: (A)Zebrafish spinal motor nerves are resorted in a plexus (DP and VP ) and connect to the muscle of the pectoral fin.
(B)Live imaging of navigation of axons along Schwann cells. Transgenic zebrafish were used to label Schwann cells.
Coordination of cohesin and DNA replication observed with purified proteins
Press release
Coordination of cohesin and DNA replication observed with purified proteins
Yasuto Murayama, Shizuko Endo, Yumiko Kurokawa, Ayako Kurita, Sanae Iwasaki, Hiroyuki Araki
Nature 2024 Jan 24 DOI:10.1038/s41586-023-07003-6
Two newly duplicated copies of genomic DNA are held together by the ring-shaped cohesin complex to ensure faithful inheritance of the genome during cell division. Cohesin mediates sister chromatid cohesion by topologically entrapping two sister DNAs during DNA replication, but how cohesion is established at the replication fork is poorly understood.
Here, we studied the interplay between cohesin and replication by reconstituting a functional replisome using purified proteins. Once DNA is encircled before replication, the cohesin ring accommodates replication in its entirety, from initiation to termination, leading to topological capture of newly synthesized DNA. This suggests that topological cohesin loading is a critical molecular prerequisite to cope with replication. Paradoxically, topological loading per se is highly rate limiting and hardly occurs under the replication-competent physiological salt concentration. This inconsistency is resolved by the replisome-associated cohesion establishment factors Chl1 helicase and Ctf4, which promote cohesin loading specifically during continuing replication. Accordingly, we found that bubble DNA, which mimics the state of DNA unwinding, induces topological cohesin loading and this is further promoted by Chl1.
Thus, we propose that cohesin converts the initial electrostatic DNA-binding mode to a topological embrace when it encounters unwound DNA structures driven by enzymatic activities including replication. Together, our results show how cohesin initially responds to replication, and provide a molecular model for the establishment of sister chromatid cohesion.
Ancient Feces Reveals Intestinal Environment of Hunters-Gatherers, Jomon People
~Metagenomic analysis of human coprolites~
Press release
Metagenomic analyses of 7000 to 5500 years old coprolites excavated from the Torihama shell-mound site in the Japanese archipelago
Luca Nishimura, Akio Tanino, Mayumi Ajimoto, Takafumi Katsumura, Motoyuki Ogawa, Kae Koganebuchi, Daisuke Waku, Masahiko Kumagai, Ryota Sugimoto, Hirofumi Nakaoka, Hiroki Oota, Ituro Inoue
PLOS ONE (2024) 19, e0295924 DOI:10.1371/journal.pone.0295924
![]() Press release (In Japanese only)
Press release (In Japanese only)
The characteristics of the intestinal environment of the Jomon people who lived in the Japanese archipelago thousands of years ago were not known.
In this study, we conducted the first metagenomic analysis in Japan using ancient DNA extracted from four Jomon-period feces fossil (coprolite) samples. Analysis of the large scale DNA data obtained by metagenomic analysis revealed genome sequences derived from bacteria and viruses that are presumed to have existed in the intestines. The results of this analysis may reflect the characteristics of the Jomon people’s intestinal environment.
We plan to conduct similar analyses on other Jomon coprolites to identify more bacteria and viruses, and to elucidate the evolution of intestinal bacteria and viruses from the Jomon period to the present day, as well as the detailed characteristics of the Jomon intestinal environment.
The research was conducted by a joint research group including Luca Nishimura (a graduate student at SOKENDAI) and Project Professor Ituro Inoue from the National Institute of Genetics, Professor Hiroki Oota from Graduate School of Science, The University of Tokyo, and Mayumi Ajimoto, a curator at the Wakasa History Museum in Fukui Prefecture.
The research results were published in the international scientific journal PLOS ONE on January 25, 2024 (JST).
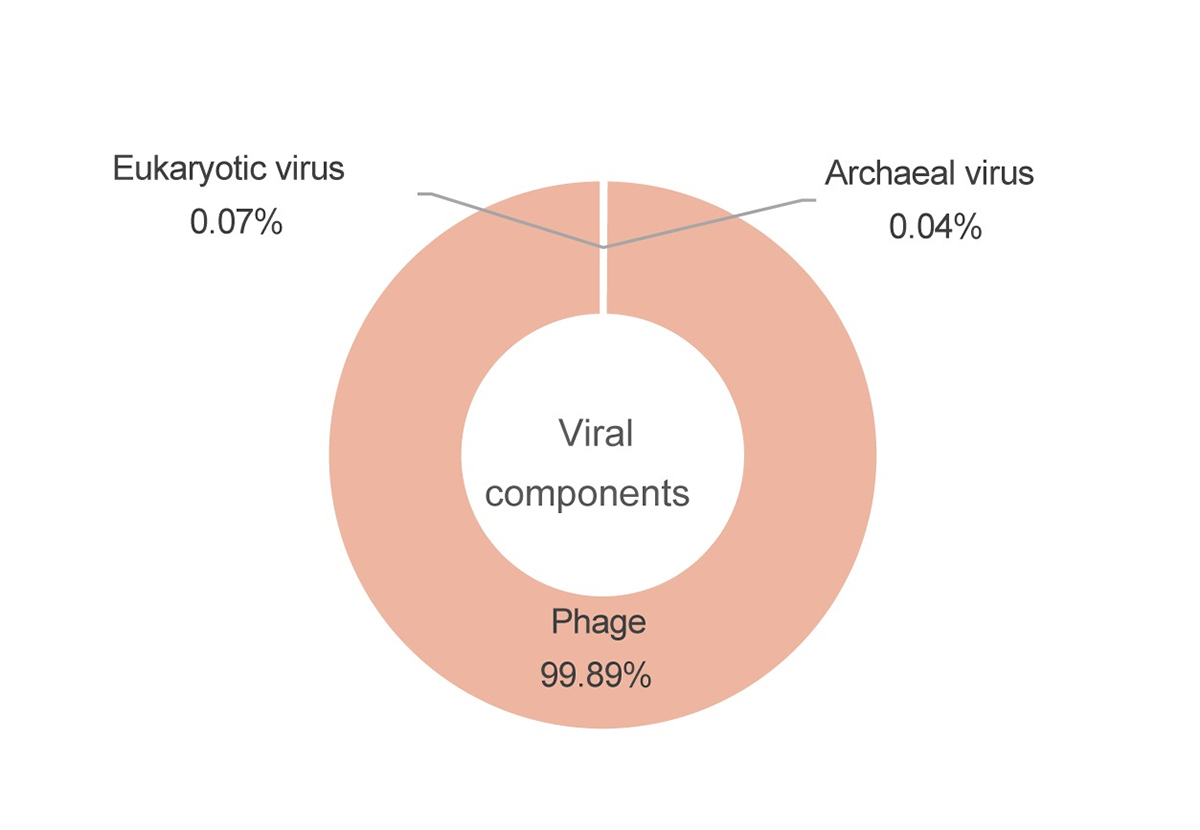
Figure: Classification of viral genome sequences detected in the Jomon coprolites.
The pie chart shows the ratio of virus from eukaryotes as 0.07%, archaea as 0.04%, and viruses infecting bacteria (phage), as 99.89%. You can see that most of the viruses are phages that
Bhim Bahadur Biswa in Mouse Genomics Resource Laboratory won the “MBSJ2023 Best Science Pitch Award” at the MSBJ2023
Bhim Bahadur Biswa(D5 student and SOKENDAI Special Researcher) in Mouse Genomics Resource Laboratory won the “MBSJ2023 Best Science Pitch Award” at the 46th Annual Meeting of the Molecular Biology Society of Japan (MBSJ2023)
- Poster Title: Changes in the gut microbiome associated with domestication of mice

ISLAM Moutushi in Molecular Cell Engineering Laboratory won the “Poster Award” at the SOKENDAI Retreat
ISLAM Moutushi in Molecular Cell Engineering Laboratory won the “Poster Award” at the SOKENDAI Retreat.
- 受賞発表タイトル: Targeted protein degradation with a single-chain antibody-based degron system.
















