Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Aoi Otsuka won the First-place Poster Award
Aoi Otsuka, D4 student and SOKENDAI Special Researcher, in Genome Dynamics Laboratory, received the First-place Poster Award at the Physical Models of Living Matters – The 3rd Symposium of Physical Biology and Biological Physics held in Taoyuan, Taiwan, on August 30th – September 1st, 2024.
Otsuka was supported by the Young Researcher Support Program of Genome modality consortium to attend the symposium.

Aoi Otsuka -san
Orientation-Independent-DIC imaging reveals that a rise in depletion attraction contributes to mitotic chromosome condensation
Press release
Orientation-Independent-DIC imaging reveals that a rise in depletion attraction contributes to mitotic chromosome condensation
Shiori Iida , Satoru Ide , Sachiko Tamura, Masaki Sasai, Tomomi Tani , Tatsuhiko Goto , *Michael Shribak , *Kazuhiro Maeshima
* Corresponding Author
Proceedings of the National Academy of Sciences (2024) 121(36), e2403153121 DOI:10.1073/pnas.240315312
![]() Press release (In Japanese only)
Press release (In Japanese only)
Mitotic chromosome condensation is an essential process to transmit replicated chromosomes into two daughter cells during cell division. To study the underlying physical principles of this process, we focused on depletion attraction/macromolecular crowding, which is a force that attracts large structures in crowded cell environments. Using special light microscopy, which can image the molecular density of cellular environments, we found that crowding around chromosomes increases during cell division. In vitro, higher concentrations of macromolecules condense chromatin and make it stiffer and more solid-like. Our results suggest that the rise in depletion attraction renders chromosomes more rigid, ensuring accurate chromosome transmission during cell division.
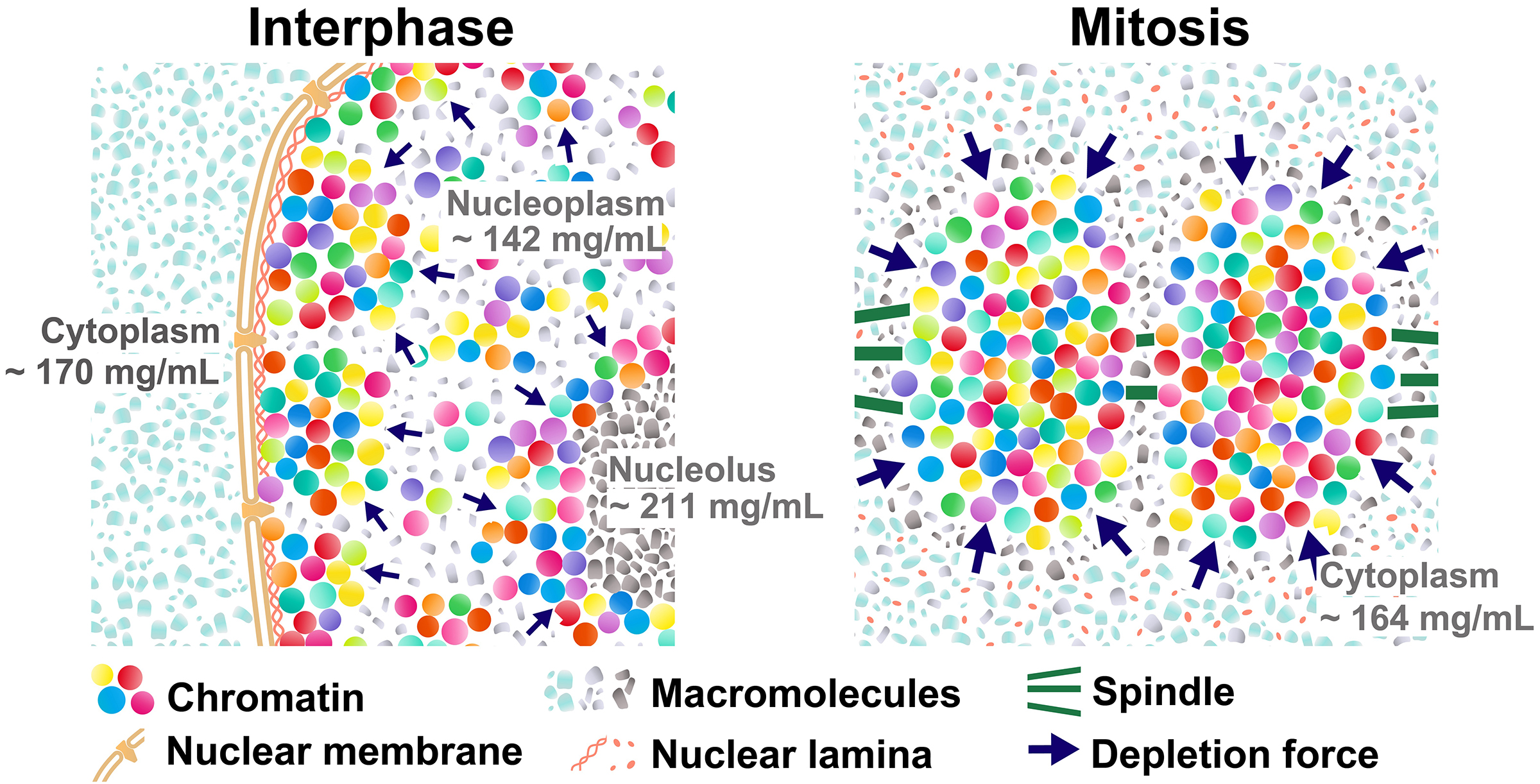
Figure: (Left) Schematic of depletion attraction in interphase live cells with soluble macromolecules in the cytoplasm (light blue), nucleoplasm (gray), and nucleolus (dark gray). The cytoplasm, nucleus, and nucleolus are compartmentalized and do not mix during interphase. Molecular densities of the nucleoplasm are lower than the cytoplasm and nucleolus. (Right) After NEBD, soluble macromolecules that were localized to the cytoplasm, nucleus, and nucleolus at interphase are now mixed. Molecular density of chromosome environment increases, making the depletion attraction stronger, and contributes to local condensation of chromosomes. These schematics are highly simplified models, and depletion attraction also works in interphase chromatin (smaller navy arrows).
Development of double-degron technology enables super-efficient and rapid degradation of target proteins.
– Cell cycle progresses to mitosis without DNA replication –
Press release
Combination of AID2 and BromoTag expands the utility of degron-based protein knockdowns
Yuki Hatoyama*, Moutushi Islam*, Adam G. Bond, Ken-ichiro Hayashi, Alessio Ciulli and Masato T. Kanemaki.
EMBO Reports (2024) Aug 23. DOI:10.1038/s44319-024-00224-4
![]() Press release (In Japanese only)
Press release (In Japanese only)
Prof. Kanemaki at the NIG and his colleagues have developed the Auxin-Inducible Degron (AID) technology, which enables rapid protein degradation for functional analysis of the proteins of interest. The group further refined it with the AID2 (Nishimura et al., Nat. Meth., 2009; Yesbolatova et al., Nat. Commun., 2020). However, they noted that some proteins remain even after degradation by AID2. In response, the group developed a double-degron technology by combining AID2 with another degron, BromoTag, achieving super-efficient and rapid degradation of target proteins. This advancement allows for the functional evaluation of proteins that were previously difficult to analyze. Additionally, they demonstrated the independent control of degradation for two different target proteins using AID2 and BromoTag.
In an application of this double-degron technology, the group successfully achieved complete inhibition of DNA replication by depleting DNA replication factors. They found that cells can progress through the cell cycle to mitosis without DNA replication, suggesting that cells may lack the system to recognize whether DNA has doubled. This finding not only demonstrates the versatility of degron technology but also offers deeper insights into the relationship between DNA replication and the cell cycle.
This study was carried out by Prof. Masato Kanemaki’s group at NIG, in collaboration with Prof. Alessio Ciulli at the University of Dundee and Prof. Kenichiro Hayashi at Okayama University of Science.
This study was supported by JSPS (21H04719, 23H04925), and JST CREST (MJCR21E6). Yuki Hatoyama and Moutushi Islam at SOKENDAI were supported by JSPS Research Fellowship (DC2) and MEXT Scholarship, respectively.
This study was published in EMBO Reports on August 23, 2024.
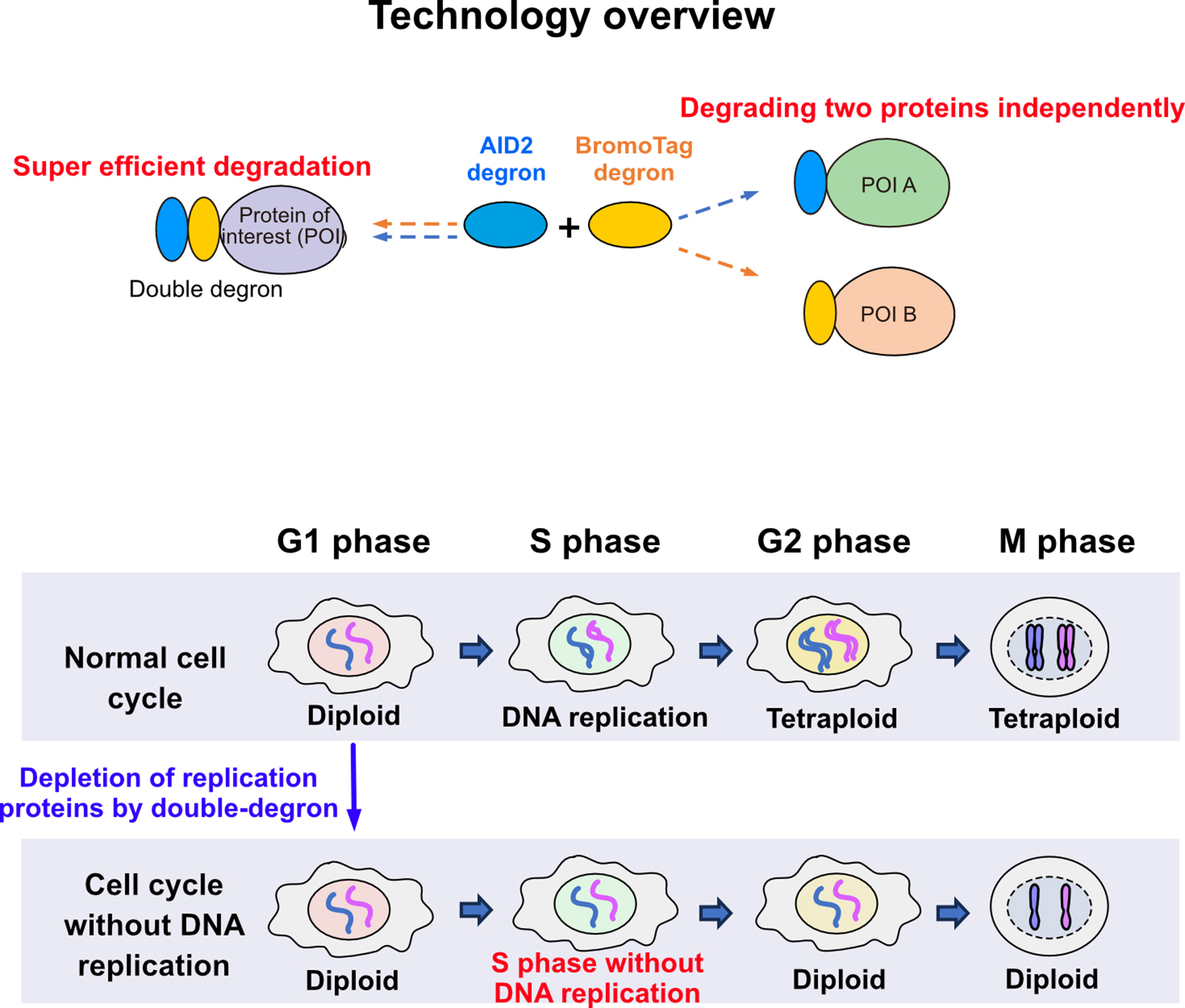
Figure: An overview of the expanded versatility achieved by combining degron technologies. The double-degron approach induced cell cycle progression to mitosis without DNA replication.
Ayjan Urazbayeva won the Best Presentation Award
Ayjan Urazbayeva, D4 SOKENDAI student and MEXT Scholar in Multiscale Sensory Structure Laboratory, received the Best Presentation Award at “Shaping the Future of Early-Career Zebrafish Researchers” Workshop in the 18th International Zebrafish Conference Conference held in Kyoto, August 17th – 21st.

Ayjan-san
Condensins and nucleosome–nucleosome interactions differentially constrain chromatin to organize mitotic chromosomes
Press release
Single-nucleosome imaging unveils that condensins and nucleosome-nucleosome interactions differentially constrain chromatin to organize mitotic chromosomes.
Kayo Hibino, Yuji Sakai, Sachiko Tamura, Masatoshi Takagi, Katsuhiko Minami, Masa A. Shimazoe, Toyoaki Natsume, Masato T. Kanemaki, Naoko Imamoto, Kazuhiro Maeshima*
* Corresponding Author
Nature Communications (2024) 15, 7152 DOI:10.1038/s41467-024-51454-y
![]() Press release (In Japanese only)
Press release (In Japanese only)
For accurate mitotic cell division, replicated chromatin must be assembled into chromosomes and faithfully segregated into daughter cells. While protein factors like condensin play key roles in this process, it is unclear how chromosome assembly proceeds as molecular events of nucleosomes in living cells and how condensins act on nucleosomes to organize chromosomes. To approach these questions, we investigate nucleosome behavior during mitosis of living human cells using single-nucleosome tracking, combined with rapid-protein depletion technology and computational modeling. Our results show that local nucleosome motion becomes increasingly constrained during mitotic chromosome assembly, which is functionally distinct from condensed apoptotic chromatin. Condensins act as molecular crosslinkers, locally constraining nucleosomes to organize chromosomes. Additionally, nucleosome-nucleosome interactions via histone tails constrain and compact whole chromosomes. Our findings elucidate the physical nature of the chromosome assembly process during mitosis.
This study was published online as open access in Nature Communications on August 21, 2024.
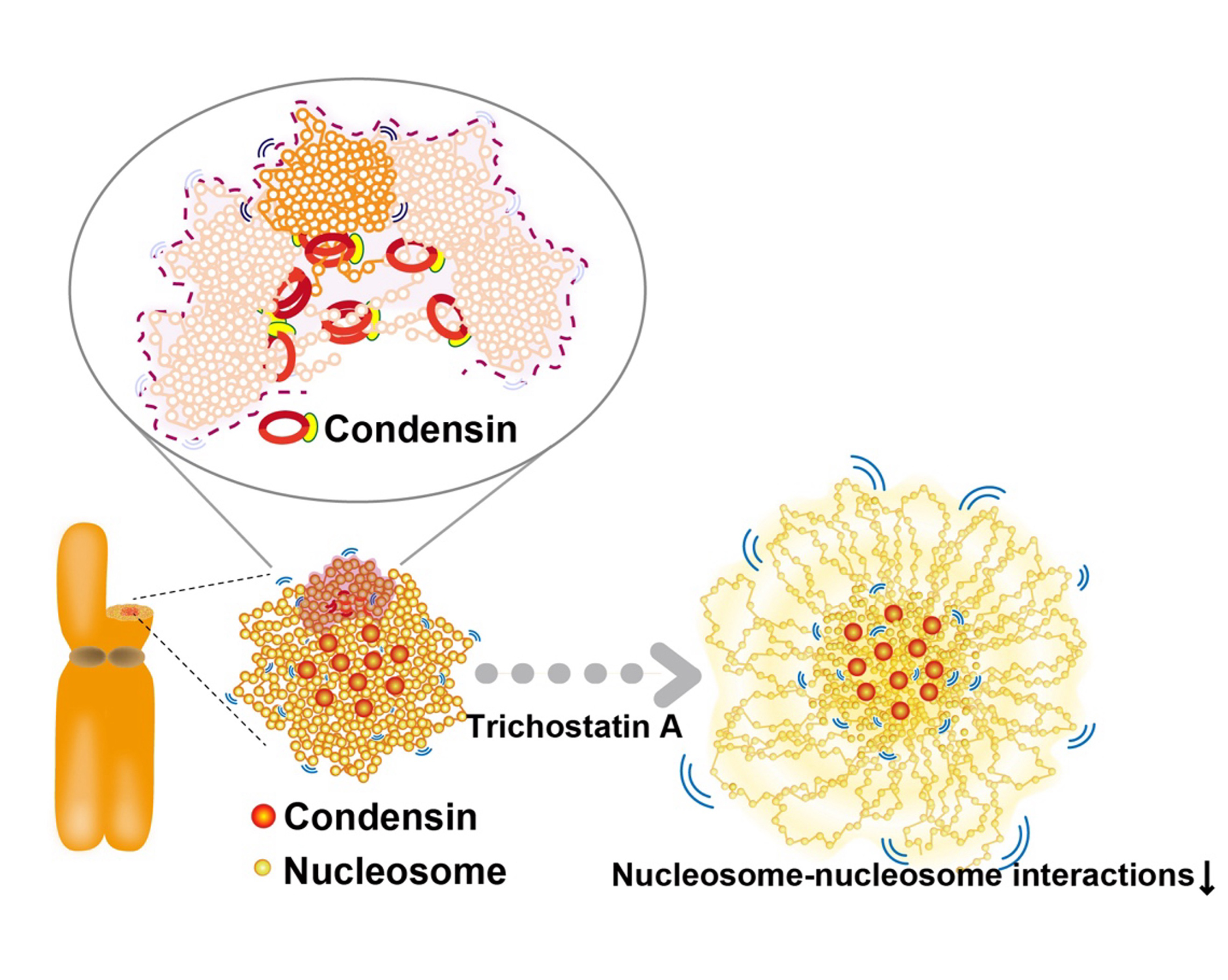
Figure: Condensins seem to act as a molecular crosslinkersto make loops. (Bottom, left) Condensins (red) locate around chromosome center. (Bottom, right) Nucleosomes around the periphery (those mostly free from condensins) in the Trichostatin A-treated chromosomes, whose nucleosome–nucleosomes interactions are weakened, are less constrained and have higher mobility than those around the axis.
Video: Single nucleosomes fluorescently labeled in a living HeLa interphase cell (left) and mitotic cell (center)(50 ms/frame). Each dot represents a single nucleosome. (Right) Calculated chromatin motion of the simulated chromosome. Chromatin beads (blue) and condensins (green and red) are shown.
Callose deficiency modulates plasmodesmata frequency in rice pollen mother cells
Nonomura Group / Plant Cytogenetics Laboratory
Koide Group / Mouse Genomics Resource Laboratory
Callose Deficiency Modulates Plasmodesmata Frequency and Extracellular Distance in Rice Pollen Mother and Tapetal cells.
Harsha Somashekar, Keiko Takanami, Yoselin Benitez-Alfonso, Akane Oishi, Rie Hiratsuka, Ken-Ichi Nonomura
Annals of Botany (2024) mcae137. DOI:10.1093/aob/mcae137
Fertilization relies on pollen mother cells able to transit from mitosis to meiosis to supply gametes. This process involves remarkable changes at the molecular, cellular and physiological levels including (but not limited to) remodelling of the cell wall. During the meiosis onset, cellulose content at the pollen mother cell walls gradually declines with the concurrent deposition of the polysaccharide callose in anther locules. We aim to understand the biological significance of cellulose-to-callose turnover in pollen mother cells walls using electron microscopic analyses of rice flowers. Our observations indicate that in wild type rice anthers, the mitosis-to-meiosis transition coincides with a gradual reduction in the number of cytoplasmic connections called plasmodesmata. A mutant in the Oryza sativa callose synthase GSL5 (Osgsl5-3), impaired in callose accumulation in premeiotic and meiotic anthers, displayed a greater reduction in plasmodesmata frequency among pollen mother cells and tapetal cells suggesting a role for callose in plasmodesmata maintenance. In addition, a significant increase in extracellular distance between pollen mother cells and impaired premeiotic cell shaping was observed in the Osgsl5-3 mutant. The results suggest that callose-to-cellulose turnover during mitosis-meiosis transition is necessary to maintain cell-to-cell connections and optimal extracellular distance among the central anther locular cells. Findings of this study contribute to our understanding of the regulatory influence of callose metabolism during meiosis initiation in flowering plants.

Figure: PD frequency is differentially regulated upon entry to meiosis and in Osgsl5-3 mutant anthers
(A) A diagram of the cross section of rice anther (left), and transmission electron microscopic images of PDs (yellow arrows) between neighbouring pollen mother cells (PMCs) in wild type (WT) (middle) and Osgsl5-3 mutant (right).
(B) Quantification of PD frequency in premeiotic interphase and early meiosis I for WT and Osgsl5-3 anthers. PD frequency was gradually decreased from premeiosis to early meiosis in WT, but significantly reduced in Osgsl5-3 mutant compared to WT.
Records of Neocaridina species in Kyushu, Japan
Kitano Group / Ecological Genetics Laboratory
Records of Neocaridina denticulata from Uku Island, N. davidi from Fukue Island, Goto Islands, and N. ikiensis from mainland Kyushu, Japan
Yusuke Fuke, Shota Kunimatsu, Jun Nakajima
Cancer (2024) 33: 47–55. DOI:10.18988/cancer.33.0_47
A freshwater shrimp, the genus Neocaridina has a land-locked life history and lives its entire life in freshwater. Two Neocaridina species are distributed in Honshu and the surrounding islands: Neocaridina denticulata and Neocaridina ikiensis, which is endemic to Iki Island. Recently, the invasive alien species Neocaridina davidi has become established in various sites in Japan, causing ecological problems.
We reported the first records of N. denticulata from Uku Island, Goto Islands, Nagasaki Prefecture, and N. ikiensis from mainland Kyushu, based on specimens. The results of mitochondrial DNA analysis suggest that these newly discovered populations are native. On the other hand, an invasive species, N. davidi was confirmed at eight sites in Fukuoka, Nagasaki, Oita, and Kumamoto prefectures. Our findings are useful for the conservation of native species and the management of invasive species.

Figure: Neocaridina denticulata collected from Uku Island. They would be a native population because they have a unique mitochondrial DNA haplotype. Photo by Jun Nakajima.
Additional records and new diagnostic traits of the freshwater prawn Macrobrachium ustulatum
Kitano Group / Ecological Genetics Laboratory
Additional records of the freshwater prawn Macrobrachium ustulatum (Crustacea: Decapoda: Palaemonidae) from the Ryukyu Islands, Japan
Yusuke Fuke, Taigi Sato, Naoto Shimizu, Naoto Inui
Cancer (2024) 33: 41–46. DOI:10.18988/cancer.33.0_41
Macrobrachium ustulatum is the sister species of Macrobrachium australe, and they have been historically confused because of their morphological similarity. The northern edge of the distribution range of both species is Japan: M. australe has been recorded from the Ryukyu Islands to a wide area of Honshu, while M. ustulatum has only two records in Japan. Therefore, the distributional records and habitat information of M. ustulatum are lacking. We report a total of 10 specimens of M. ustulatum collected from three localities in the Ryukyu Islands. The environments in which these specimens were collected suggest that the habitat of this species is a weakly flowing environment in the middle reach of a river. This differs slightly from M. australe, which prefers stagnant environments. Furthermore, we have succeeded in proposing new distinguishing characteristics for both species.

Figure: Macrobrachium ustulatum (top) and M. australe (bottom). Photo by the authors.
Confusion between Caridina laoagensis and Caridina tupaia in Japan
Kitano Group / Ecological Genetics Laboratory
Confusion between Caridina laoagensis and Caridina tupaia in Japan
Yusuke Fuke & Tomoaki Maruyama
Cancer (2024) 33: 15–23. DOI:10.18988/cancer.33.0_15
The morphological characters of the freshwater shrimp called “Ryugu-hime-ebi” in Japan are not always consistent with those of Caridina laoagensis, which has been given its Japanese name. Recently, a cryptic species of C. weberi species group that includes C. laoagensis was newly described as Caridina tupaia. We aimed to resolve the historical confusion between C. laoagensis and C. tupaia in Japan. For DNA barcoding and morphological examination, we recognized the two taxa in “Ryugu-hime-ebi,” i.e. C. laoagensis and C. tupaia. Based on some diagnostic characters, we reviewed previous records and found that the taxon that was given the Japanese name “Ryugu-hime-ebi” was most likely C. tupaia. We proposed to give the standard Japanese name “Ryugu-hime-ebi” to C. tupaia and the new standard Japanese name “Un-mon-hime-ebi” to C. laoagensis.
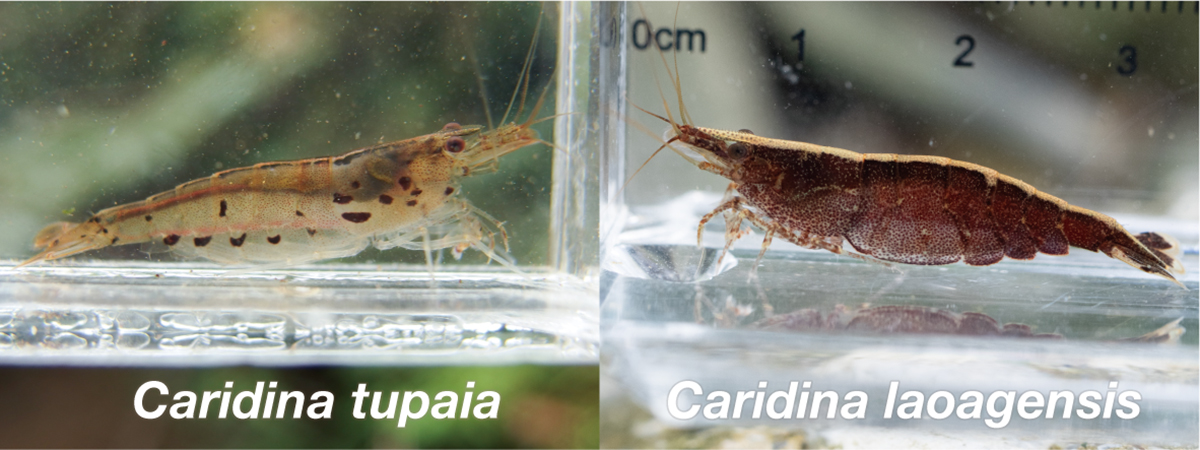
Figure: Caridina tupaia and C. laoagensis. Photo by the authors.
Adilgazy Semeigazin won the Poster Award and Travel Award
Adilgazy Semeigazin, D4 student and MEXT Scholar in Genome Dynamics Laboratory, received the Second-place Poster Award at the FASEB Research Conference ”Biology of Acetylation in Health and Disease“ held in Rome, Italy, on August 4th – 8th. He also received the Travel Award.
Semeigazin was also supported by SOKENDAI Student Dispatch Program to attend the conference.

Semeigazin-san
Cytoplasmic RNA granule required for regulation of meiosis transition in rice
Nonomura Group / Plant Cytogenetics Laboratory
Impact of protein domains on the MEL2 granule, a cytoplasmic ribonucleoprotein complex maintaining faithful meiosis progression in rice.
Manaki Mimura, Seijiro Ono, Harsha Somashekar, Ken-Ichi Nonomura
New Phytologist 2024 Jul 24. DOI:10.1111/nph.19968
Cytoplasmic ribonucleoprotein (RNP) granules are membraneless structures composed of various RNAs and proteins that play important roles in post-transcriptional regulation. While RNP granules are known to regulate the meiotic entry in some organisms, little is known about their roles in plants.
In this study, we observed the cytoplasmic granular structures of rice RNA-binding protein MEL2, which contributes to the control of meiotic entry timing, in leaf protoplasts and spore mother cells. Our results indicated that MEL2 granules colocalized with processing body and stress granule factors. The maintenance of granule properties modulated by LOTUS domain and the intrinsically disordered region (IDR) is essential for proper MEL2 function in meiosis progression (Figure). MEL2-like proteins widely found in plant kingdom conserved LOTUS domain followed by the IDR despite their diverse domain structures, suggesting the functional conservation of these domains among plant species.
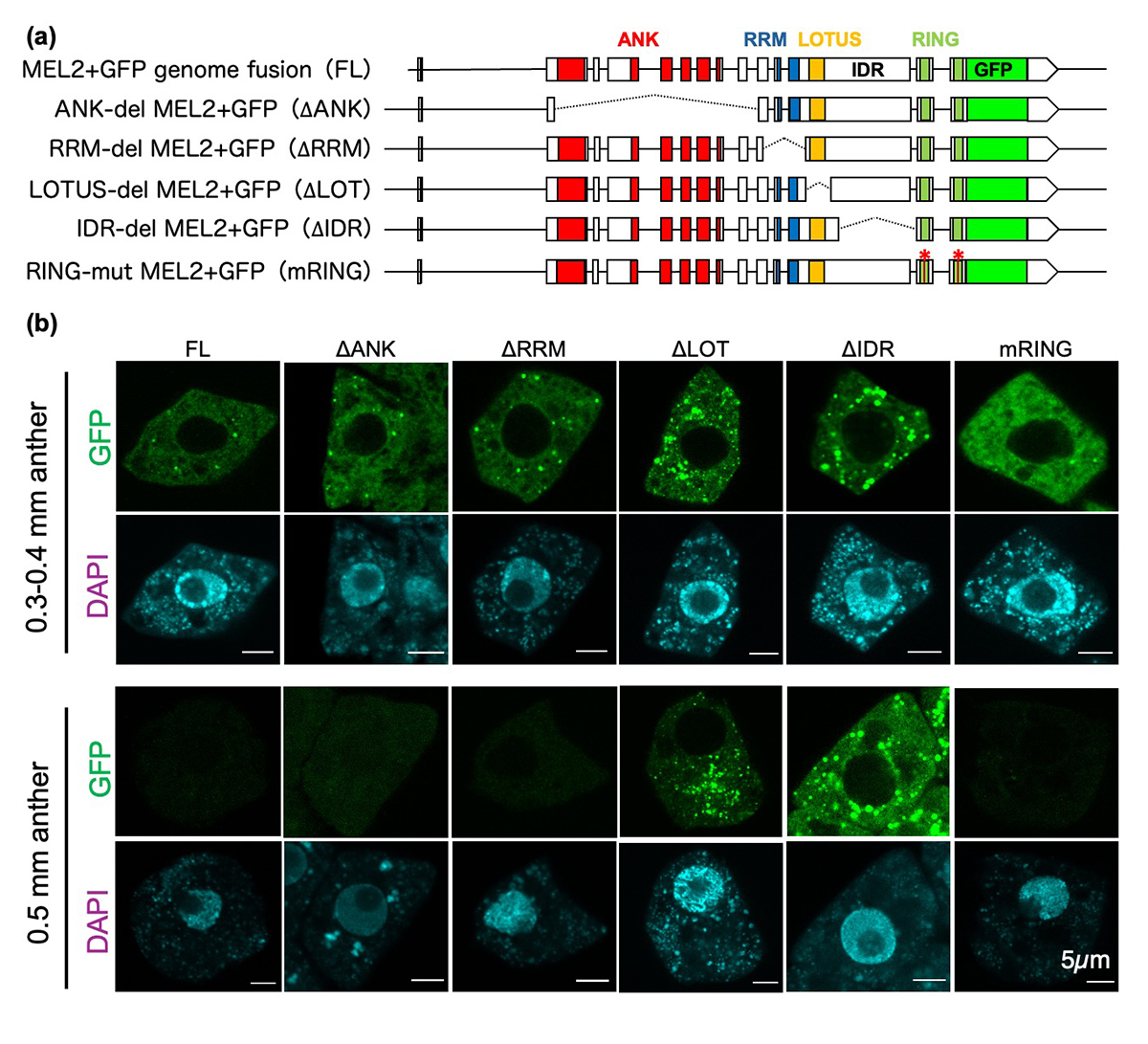
Figure: Roles of MEL2 functional domains in granule formation
(a) Genomic fusion constructs of MEL2 and GFP genes, and its derivatives with deletions (dashed lines) or mutations (asterisks) in MEL2 domains.
(b) MEL2 granule formation in spore mother cells in rice anthers transformed with the constructs in (a). The wildtype MEL2 (FL) formed granules (green) at premeiosis (0.3-0.4mm anthers), and disappeared by meiosis (0.5mm anthers). The mRING failed granule formation, and the granules of ∆LOTUS and ∆IDR were aberrantly carried over to meiocytes, all resulting in defective meiosis and pollen development.
The First Dimerization-Dependent Protein Knockdown Method in Photosynthetic Organisms
Miyagishima Group / Symbiosis and Cell Evolution Laboratory
Development of a rapamycin-inducible protein-knockdown system in the unicellular red alga Cyanidioschyzon merolae.
Takayuki Fujiwara, Shunsuke Hirooka, Shota Yamashita, Fumi Yagisawa and Shin-ya Miyagishima
Plant Physiology (2024) kiae316 DOI:10.1093/plphys/kiae316
An inducible protein-knockdown system is highly effective for investigating the functions of proteins and mechanisms essential for the survival and growth of organisms. However, this technique is not available in photosynthetic eukaryotes. The unicellular red alga Cyanidioschyzon merolae possesses a very simple cellular and genomic architecture and is genetically tractable but lacks RNA interference machinery. In this study, we developed a protein-knockdown system in this alga. The constitutive system utilizes the destabilizing activity of the FK506-binding protein 12 (FKBP12)-rapamycin-binding (FRB) domain of human target of rapamycin kinase or its derivatives to knock down target proteins. In the inducible system, rapamycin treatment induces the heterodimerization of the human FRB domain fused to the target proteins with the human FKBP fused to S-phase kinase associated protein 1 or Cullin 1, subunits of the SCF E3 ubiquitin ligase. This results in the rapid degradation of the target proteins through the ubiquitin-proteasome pathway. With this system, we successfully degraded endogenous essential proteins such as the chloroplast division protein dynamin-related protein 5B and E2 transcription factor, a regulator of the G1/S transition, within 2 to 3 h after rapamycin administration, enabling the assessment of resulting phenotypes. This rapamycin-inducible protein-knockdown system contributes to the functional analysis of genes whose disruption leads to lethality.
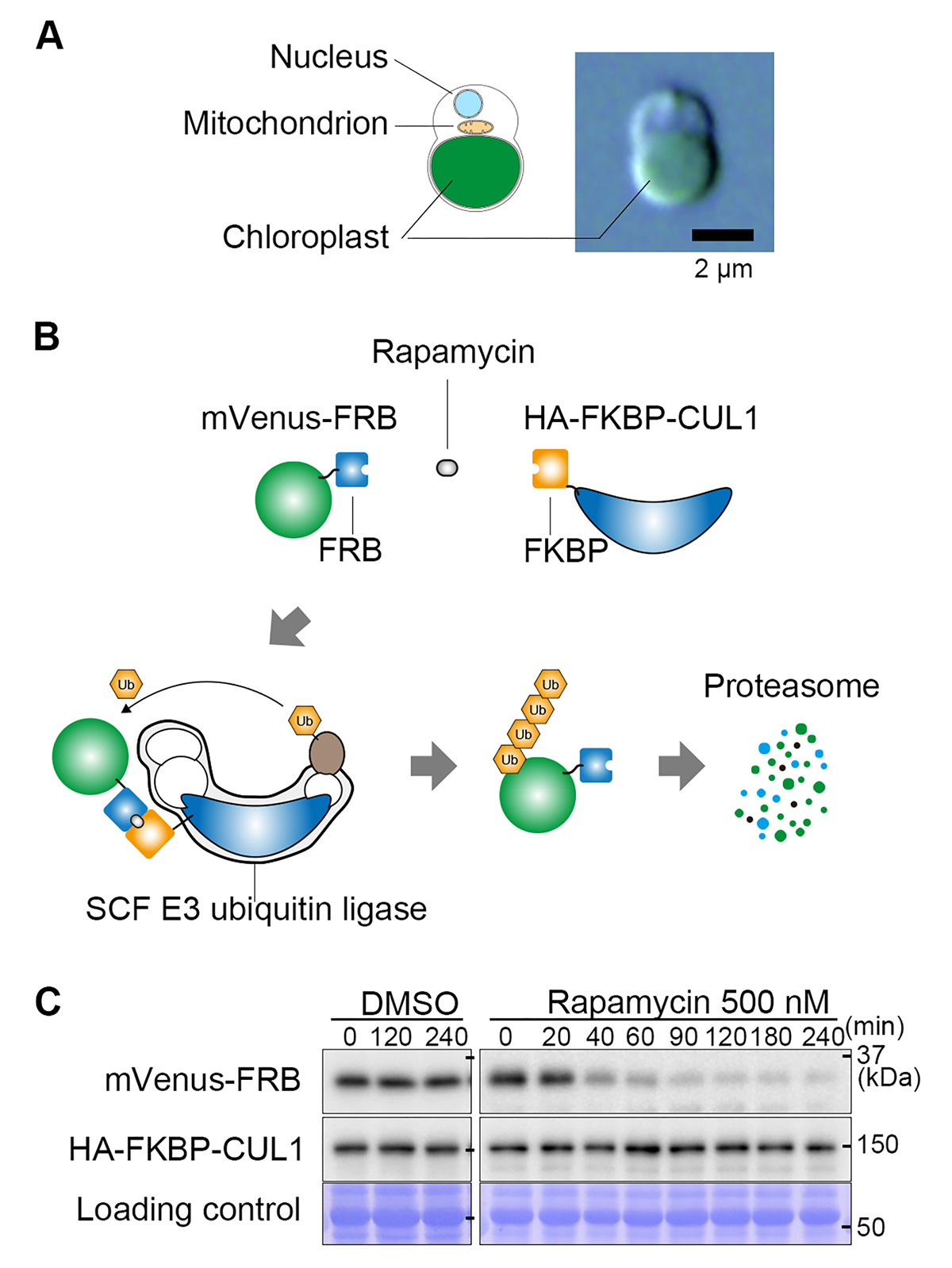
Figure: A rapamycin-inducible protein knockdown system.
(A) The cellular structure of the unicellular red alga Cyanidioschyzon merolae and a differential interference contrast microscopy image.
(B) Mechanism of the rapamycin-inducible protein knockdown system. FRB tags and FKBP proteins form a heterodimer in the presence of rapamycin. The target protein fused with an FRB tag (in this figure, the fluorescent protein mVenus) and the CUL1 subunit of ubiquitin ligase fused with FKBP bind in the presence of rapamycin, resulting in the ubiquitination of FRB-mVenus. The ubiquitinated FRB-mVenus is rapidly degraded by the proteasome.
(C) Immunoblot analysis showing the decrease in mVenus-FRB protein level after the addition of rapamycin.
Summer Holiday (Aug. 15-16)
NIG will be closed from August 15 to August 16, 2024 for summer holiday.
Thank you for your understanding and cooperation.
NIG website service will be stopped several times. July 11 Thu, 00:00 – July 11 Thu, 08:00.
Due to the facility maintenance, the NIG website service will be stopped several times on the following time.
Thank you for your understanding and cooperation.
Time and Date: July 11 Thu, 00:00 – July 11 Thu, 08:00.
以下の期日において、遺伝研公式ウェブサイトの断続的な停止を伴う保守作業が予定されております。
皆様のご理解とご協力をお願いいたします。
停止日時: 7月11日(木) 00:00~7月11日(木) 08:00
理由: 保守作業のため
Problems in Sending and Receiving NIG E-mails between 12:45-13:45 on July 4
Due to the test of the NIG mail system, NIG account mails may not be able to send or receive between 12:45-13:45 on July 4.
We apologize for any inconvenience and appreciate your understanding and cooperation.
Where are the replication initiation zones located in mammalian genome? (Review paper)
Kanemaki Group / Molecular Cell Engineering Laboratory
Replication initiation sites and zones in the mammalian genome: Where are they located and how are they defined?
Xiaoxuan Zhu, Masato T. Kanemaki*
*Corresponding author
DNA Repiar (2024) 141, 103713 DOI:10.1016/j.dnarep.2024.103713
The question of how genomic DNA is replicated is one of the key themes in molecular biology, as abnormalities in DNA replication are related to genome instability, genetic disorders, cell death, and cancer. Therefore, DNA replication has been extensively studied over the years using Escherichia coli and budding yeast as model organisms. In these unicellular models, DNA replication starts from genomic regions with consensus DNA sequences. On the other hand, in mammalian cells, including humans, consensus sequences appear to be absent, and it is still not well understood where DNA replication begins and how are they defined in the genome. Hence, this review paper compiles and compares the results of large-scale high-throughput sequencing-based explorations for mapping initiation sites/zones reported so far. Furthermore, it highlights the similarities and differences in the DNA replication mechanisms between budding yeast and mammalian cells, presents a hypothetical mechanism for how replication initiation zones are defined in mammalian cells, and discusses the future directions and challenges in DNA replication research.
The first author, Dr Xiaoxuan Zhu, has been supported as an NIG post-doctoral fellow.
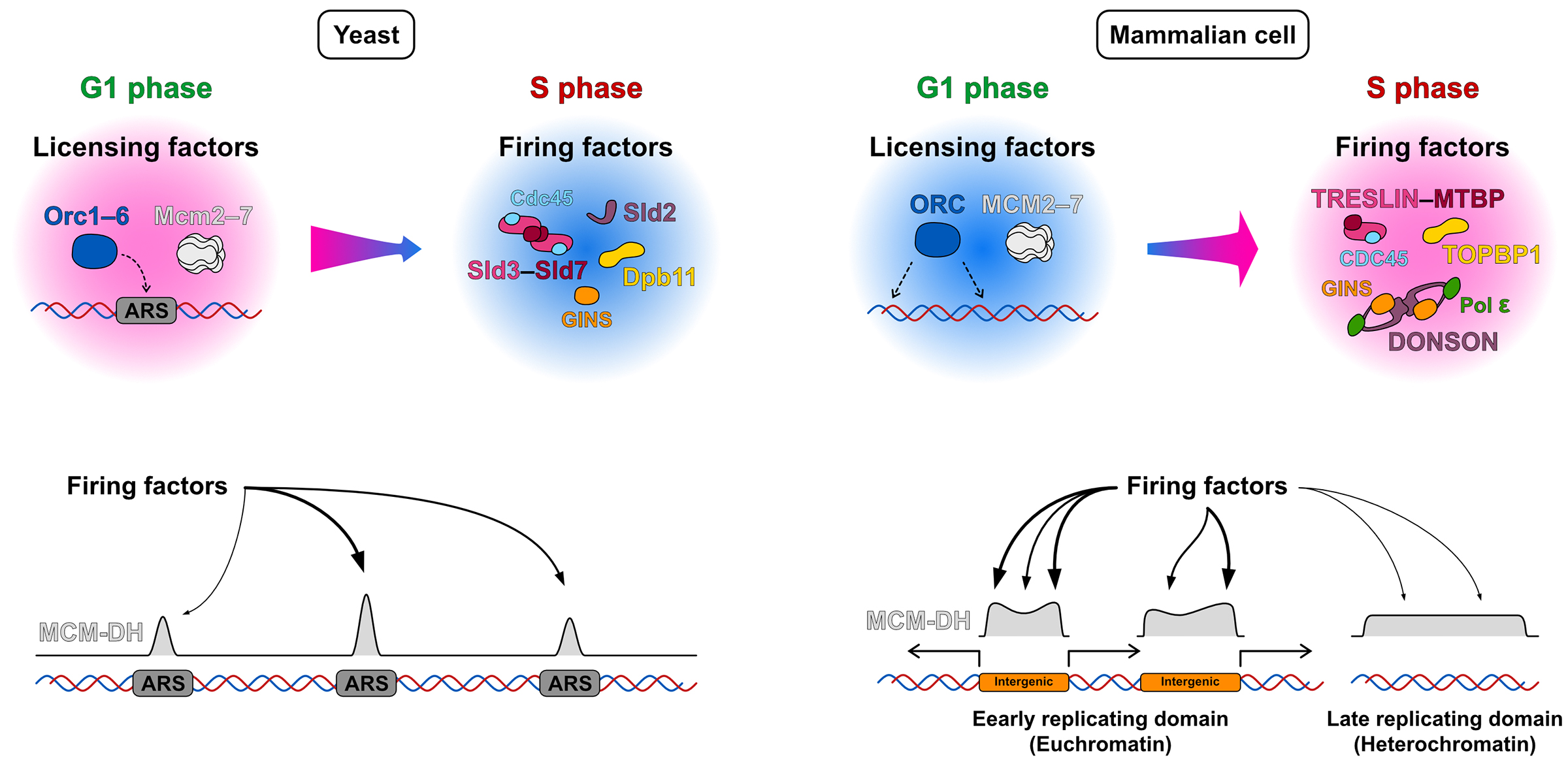
Figure: Differences in the mechanisms of replication initiation sites/zones selection between budding yeast and mammalian cells: In yeast, licensing factors accumulate at ARS (autonomously replicating sequence) sites containing a consensus sequence, leading to replication initiation at these ARS sites. In contrast, in mammalian cells, licensing factors are widely distributed across the genome. It is thought that the subsequent accumulation of firing factors at specific locations is crucial for forming initiation zones.
YABBY and diverged KNOX1 genes shape nodes and internodes in the stem
Press release
Nonomura Group / Plant Cytogenetics Laboratory
Technical Section/ Cell Architecture Laboratory
YABBY and diverged KNOX1 genes shape nodes and internodes in the stem
Katsutoshi Tsuda*, Akiteru Maeno, Ayako Otake, Kae Kato, Wakana Tanaka, Ken-Ichiro Hibara, and Ken-Ichi Nonomura
* Corresponding author
Science (2024) 384, 1241-1247 DOI:10.1126/science.adn6748
![]() Press release (In Japanese only)
Press release (In Japanese only)
Plant stems comprise nodes and internodes that specialize in solute exchange and elongation. However, their boundaries are not well defined, and how these basic units arise remains elusive. In rice with clear nodes and internodes, we found that one subclade of class I knotted1-like homeobox (KNOX1) genes for shoot meristem indeterminacy restricts node differentiation and allows internode formation by repressing YABBY genes for leaf development and genes from another node-specific KNOX1 subclade. YABBYs promote nodal vascular differentiation and limit stem elongation. YABBY and node-specific KNOX1 genes specify the pulvinus, which further elaborates the nodal structure for gravitropism. Notably, this KNOX1 subclade organization is specific to seed plants. We propose that nodes and internodes are distinct domains specified by YABBY–KNOX1 cross-regulation that diverged in early seed plants.
Source: Katsutoshi Tsuda et al., Science (2024) 384, 1241-1247
A novel gene‐trap line reveals the dynamic patterns and essential roles of cysteine and glycine‐rich protein 3 in zebrafish heart development and regeneration
Kawakami Group / Laboratory of Molecular and Developmental Biology
A novel gene‐trap line reveals the dynamic patterns and essential roles of cysteine and glycine‐rich protein 3 in zebrafish heart development and regeneration.
Shuzhang Liang, Yating Zhou, Yue Chang, Jiayi Li, Min Zhang, Peng Gao, Qi Li, Hong Yu, Koichi Kawakami, Jinmin Ma, and Ruilin Zhang.
Cellular and Molecular Life Sciences (2024) 81, 158 DOI:10.1007/s00018-024-05189-
CSRP3/MLP, a key regulator of striated muscle function, have been linked to hypertrophic cardiomyopathy (HCM) and dilated cardiomyopathy (DCM) in patients. However, the roles of CSRP3 in heart development and regeneration are not completely understood. In this study, we characterized a novel zebrafish gene-trap line, gSAIzGFFM218A, which harbors an insertion in the csrp3 genomic locus. We discovered that csrp3 is specifically expressed in larval ventricular cardiomyocytes (CMs) and that csrp3 deficiency leads to excessive trabeculation, a common feature of CSRP3-related HCM and DCM. We further revealed that csrp3 expression increased in response to different cardiac injuries and was regulated by several signaling pathways vital for heart regeneration. Csrp3 deficiency impeded heart regeneration by impairing CM dedifferentiation, hindering sarcomere reassembly, and reducing CM proliferation while aggravating apoptosis. Csrp3 overexpression promoted CM proliferation after injury and ameliorated the impairment of ventricle regeneration caused by pharmacological inhibition of multiple signaling pathways. Our study highlights the critical role of Csrp3 in both heart development and regeneration, and provides a valuable animal model for further functional exploration that will shed light on the molecular pathogenesis of CSRP3-related human cardiac diseases.
This study was conducted as a collaborative research between the Zhang Lab at Wuhan University and the Kawakami Laboratory. This study was supported by NIG-JOINT (2015 Collaborative Research A1-8).
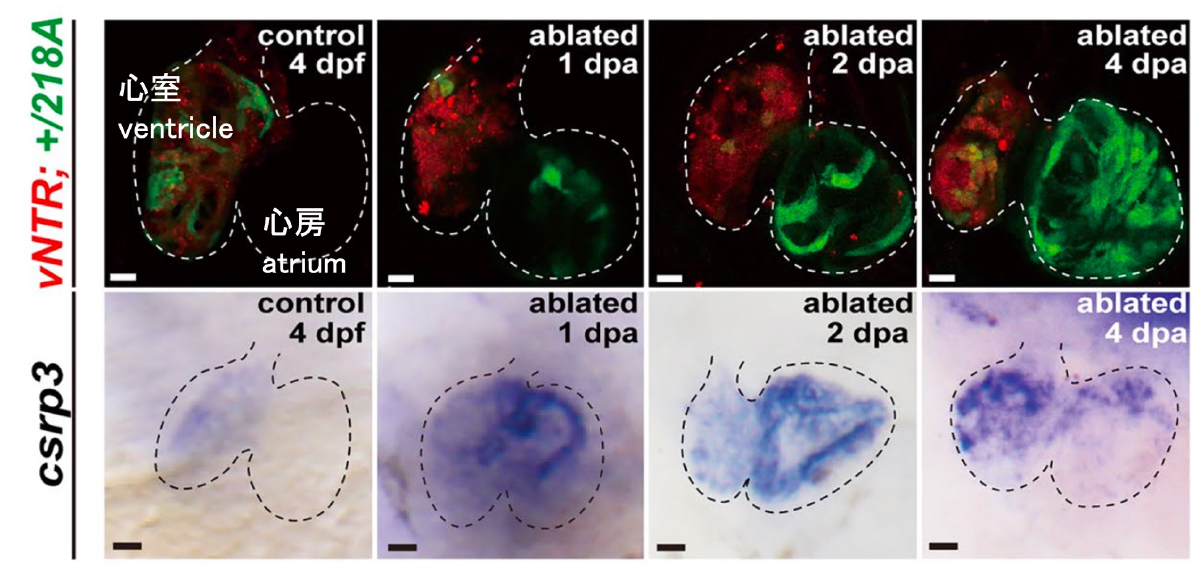
Figure: Induction of csrp3 gene expression in the atrium of the heart after ventricular cardiomyocyte ablation
(Red) Expression of mCherry-NTR in the ventricular cardiomyocytes. Cells were ablated by addition of MTZ.
(Green) Induction of csrp3 gene expression in the heart after ventricular cardiomyocyte ablation.
A conserved gene regulatory network tells neurons to send axons across the midline
Hirata Group / Brain Function Laboratory
A conserved gene regulatory network tells neurons to send axons across the midline
Aki Masuda, Kazuhiko Nishida, Rieko Ajima, Yumiko Saga, Marah Bakhtan, Avihu Klar, Tatsumi Hirata, Yan Zhu*
* Corresponding author
Science Advances (2024) 10, eadk2149 DOI:10.1126/Sciadv.adk2149
All neurons can be divided into two types: those that send their axons across the midline (commissural), and those that do not (ipsilateral). This binary division takes place broadly across heterogenous classes of neurons. To date, very little is known of the nature of the genetic program(s) that drives this axon laterality-based division. In this study, we have uncovered an evolutionarily conserved gene regulatory mechanism, involving a pair of basic helix-loop-helix transcription factors Nhlh1/2, that is essential for the specification of all floor plate-crossing commissural neurons from the spinal cord to the midbrain. We have further shown that this global mechanism is regulated regionally by neuron class determinants to achieve specificities of the commissural and ipsilateral division. Our work has shed light on a molecular strategy in which an interplay between a globally-acting program and the lineage-based regional programs controls a common trait shared across heterogenous neuronal classes.
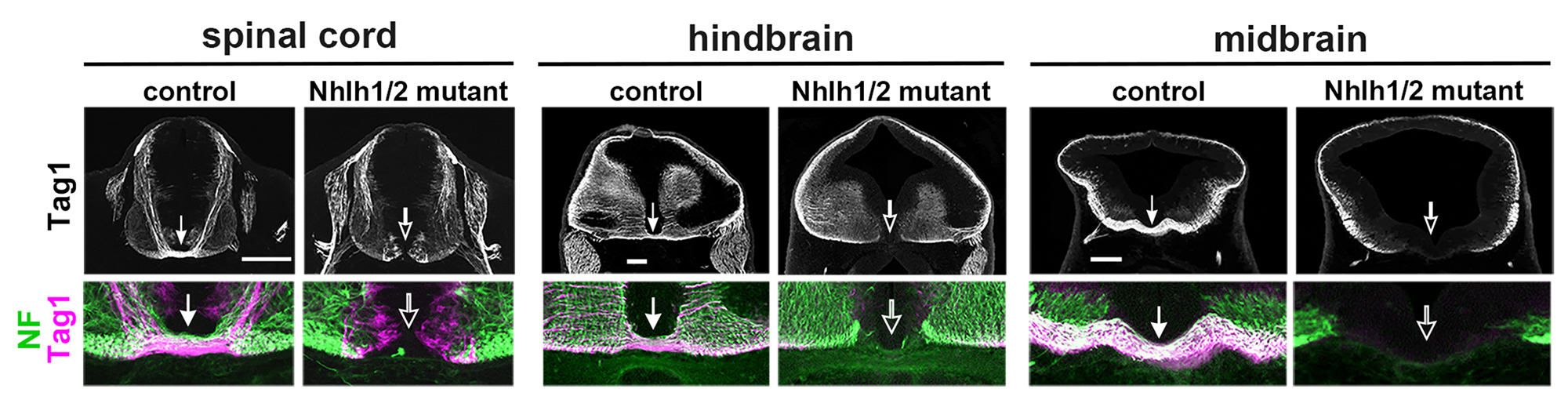
Figure1: Complete lack of axons crossing the midline of the neural tube in Nhlh1/2 double mutant mice.
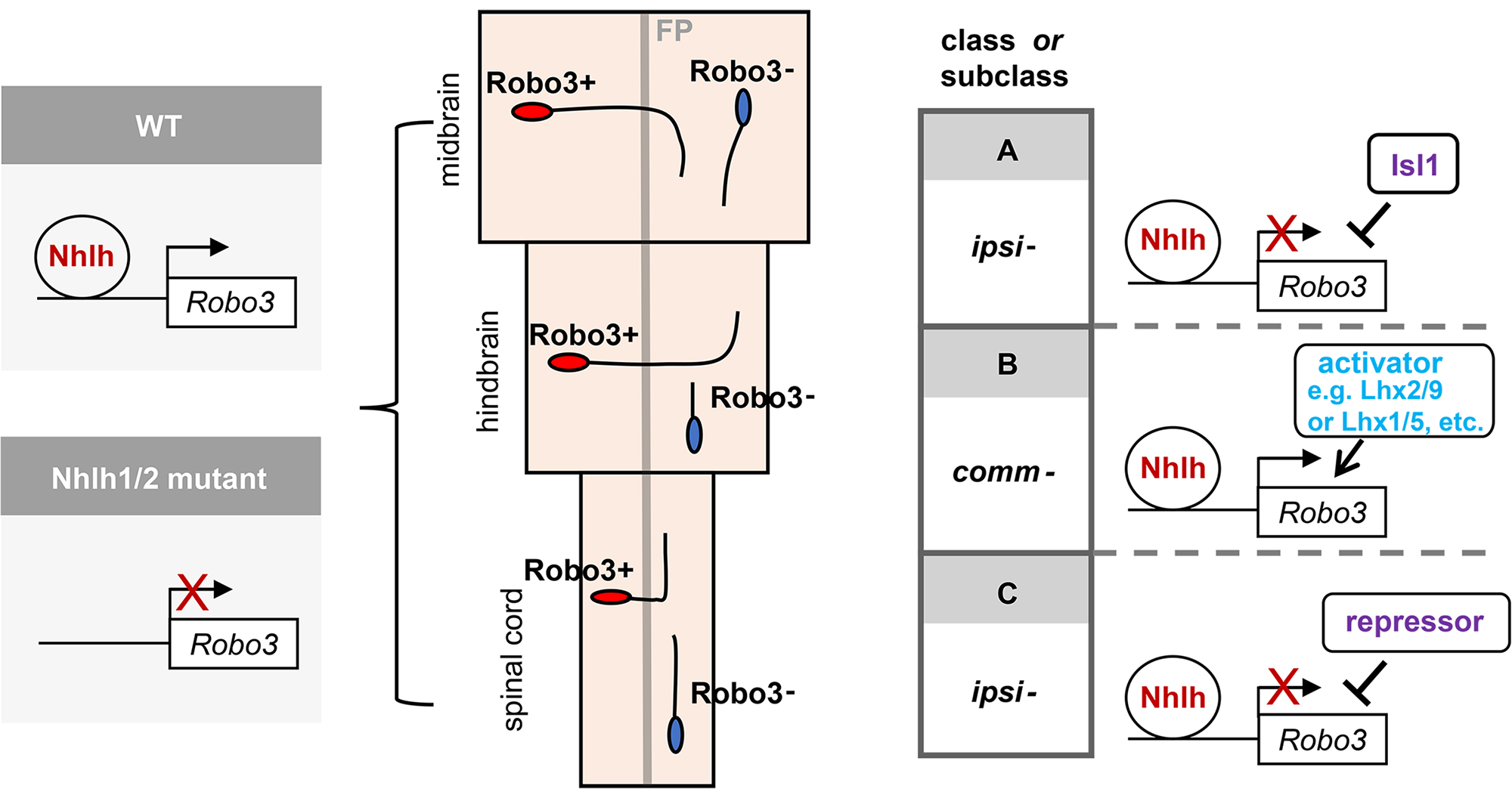
Figure2: Transcription factors Nhlh1/2 bind to an enhancer region of the guidance receptor Robo3 and activate its transcription. Depletion of Nhlh1/2 results in a large reduction of Robo3 expression and consequentially failure of axons crossing the midline of the neural tube. The activity of Nhlh1/2 is refined by neuronal class or subclass specific regulators, such as Isl1, and possibly Lhx family members.















