Kitano Group • Ecological Genetics Laboratory
Genetics of adaptive radiation
Faculty
Research Summary
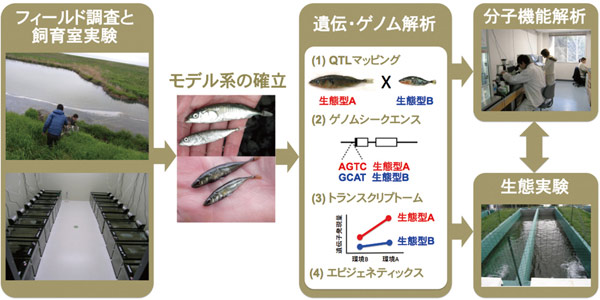
Our research goal is to understand the molecular mechanisms underlying the evolution of biodiversity. Although many genes important for animal development and behavior have been identified in model organisms, little is known about the molecular mechanisms underlying naturally occurring phenotypic variation important for adaptation and speciation in wild populations. Furthermore, little is known about how newly evolved alleles important for adaptation and speciation spread within natural populations. To understand these ecological and genetic mechanisms, we mainly use stickleback fishes as a model. Our research takes an integrative approach across diverse disciplines.
Selected Publications
Kitano J, Ansai S, Fujimoto S, Kakioka R, Sato M, Mandagi IF, Sumarto BKA, Yamahira K. A Cryptic Sex-Linked Locus Revealed by the Elimination of a Master Sex-Determining Locus in Medaka Fish. Am Nat. 2023 Aug;202(2):231-240.
Yoshida K, Kitano J. Tempo and mode in karyotype evolution revealed by a probabilistic model incorporating both chromosome number and morphology. PLoS Genet. 2021 Apr 16;17(4):e1009502.
Ansai S, Mochida K, Fujimoto S, Mokodongan DF, Sumarto BKA, Masengi KWA, Hadiaty RK, Nagano AJ, Toyoda A, Naruse K, Yamahira K, Kitano J. Genome editing reveals fitness effects of a gene for sexual dichromatism in Sulawesian fishes. Nat Commun. 2021 Mar 1;12(1):1350.
Ravinet M, Kume M, Ishikawa A, Kitano J. Patterns of genomic divergence and introgression between Japanese stickleback species with overlapping breeding habitats. J Evol Biol. 2021 Jan;34(1):114-127.

















