Koide Group • Mouse Genomics Resource Laboratory
Behavioral genetics using wild-derived mouse strains
Faculty
Research Summary
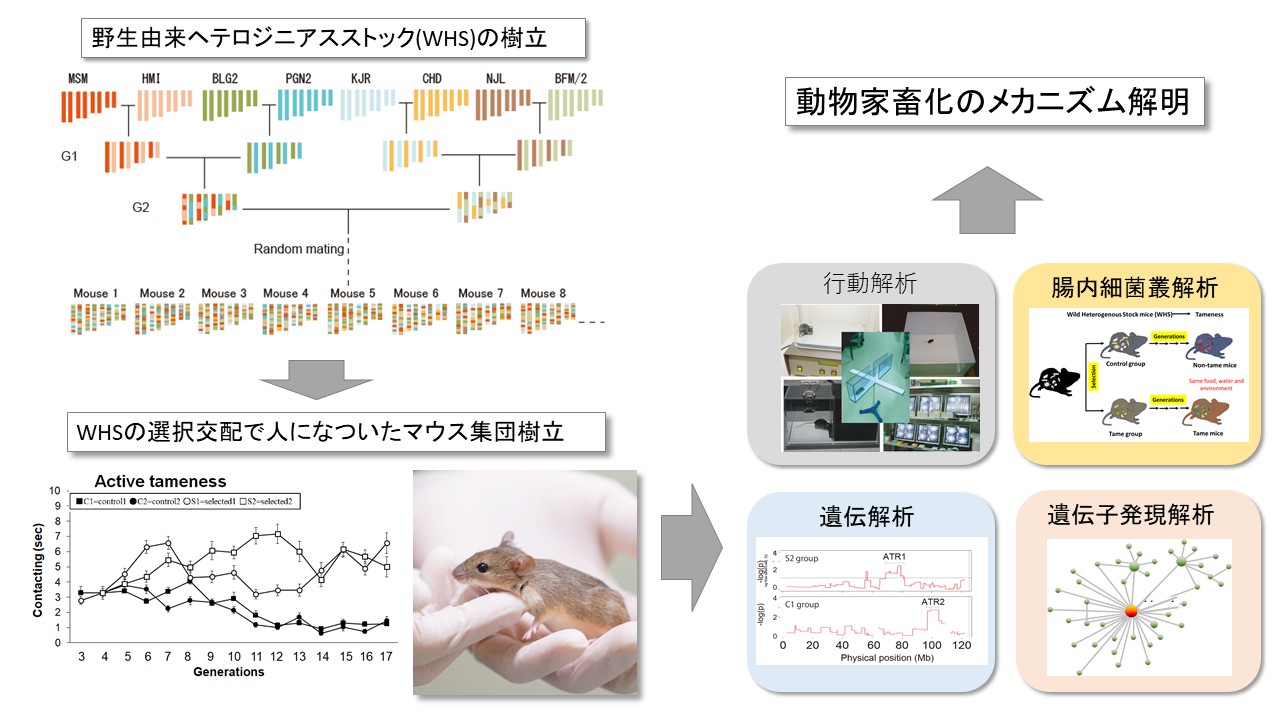
The genetic basis for individual differences in complex traits is still unclear. In order to clarify the mechanisms related to behavioral diversity, we are using a series of wild-derived mouse strains. Wild derived strains exhibit a prominent degree of wildness and phenotypic diversity among them. We are also developing efficient genome editing methodologies in rodents with CRISPR/Cas9. We are identifying genes related to behavioral diversity using these tools, and are aiming to understand the role of these genes in the molecular, cellular, and neural mechanisms that underlie this behavioral diversity.
Selected Publications
Nakamura M, Nomoto K, Mogi K, Koide T, Kikusui T. Visual and olfactory signals of conspecifics induce emotional contagion in mice. Proc Biol Sci. 2024 Dec;291(2036):20241815.
Niimura Y, Biswa BB, Kishida T, Toyoda A, Fujiwara K, Ito M, Touhara K, Inoue-Murayama M, Jenkins SH, Adenyo C, Kayang BB, Koide T. Synchronized Expansion and Contraction of Olfactory, Vomeronasal, and Taste Receptor Gene Families in Hystricomorph Rodents. Mol Biol Evol. 2024 Apr 2;41(4):msae071.
Venkatachalam B, Biswa BB, Nagayama H, Koide T. Association of tameness and sociability but no sign of domestication syndrome in mice selectively bred for active tameness. Genes Brain Behav. 2024 Feb;23(1):e12887.
Takanami K, Kuroiwa M, Ishikawa R, Imai Y, Oishi A, Hashino M, Shimoda Y, Sakamoto H, Koide T. Function of gastrin-releasing peptide receptors in ocular itch transmission in the mouse trigeminal sensory system. Front Mol Neurosci. 2023 Nov 30;16:1280024.
















