Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Dr. Huong Ta received the Best Papers Award
Dr. Huong Ta, a project postdoc in Plant Cytogenetics Lab./Nonomura Lab., received the Best Papers Award 2024 in the 96th Annual Meeting of the Genetics Society of Japan.
・Title: AGO4a contributes to the epigenetic shaping of meiotic DNA methylome in rice
・The Winners List

Dr. Huong Ta (left) with Prof Nonomura
Neuronal fate resulting from indirect neurogenesis in the mouse neocortex
Hirata Group / Brain Function Laboratory
Neuronal fate resulting from indirect neurogenesis in the mouse neocortex
Hatanaka Y, Yamada K, Eritate T, Kawaguchi Y, Hirata T.
Cerebral Cortex Volume 34, Issue 11, November 2024, bhae439 DOI:10.1093/cercor/bhae439
The mammalian neocortex is characterized by a six-layered structure, with each layer containing neurons that perform distinct functions. These neurons are derived from neural stem cells, known as radial glial cells (RGCs), located in the ventricular zone. During neurogenesis, RGCs divide by two distinct modes: direct neurogenesis, where RGCs generate neurons directly, and indirect neurogenesis, where RGCs first produce intermediate neural progenitors (INPs) that subsequently differentiate into neurons. Although these differentiation modes have been demonstrated through in vitro imaging studies, their contributions to cortical structure formation remain poorly understood.
We developed a method to identify neurons generated via indirect neurogenesis to address this issue. First, we demonstrated that tamoxifen administration to Neurog2CreER mice induces recombination in cells committed to becoming neurons, including INPs. Second, we distinguished neurons generated via indirect neurogenesis by allowing them to incorporate a thymidine analog. Using this approach, we found that indirect neurogenesis is prominent during the early stages of cortical neurogenesis and that both neurogenesis modes generate similar neuronal types in the same temporal sequence. Moreover, by crossing Neurog2CreER mice with MADM mice, we demonstrated that INPs undergo a single division to produce two neurons exhibiting similar morphology.
These findings indicate that direct and indirect neurogenesis cooperatively generate a diverse array of neuronal types in a temporally coordinated manner, and their progeny populate together to form a coherent cortical structure. This study also provides important insights into the mechanisms underlying neocortical expansion during evolution.
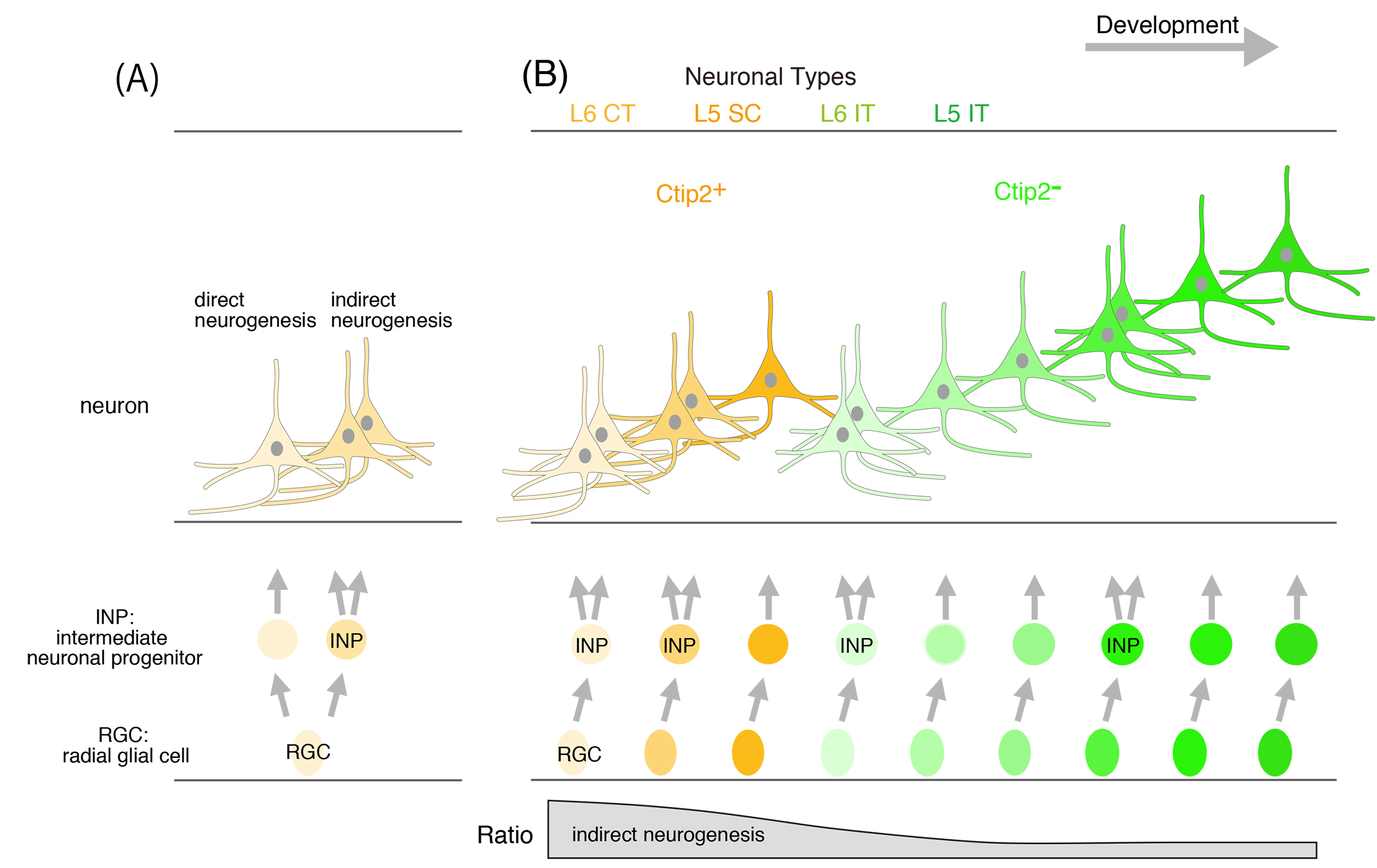
Figure: (A) Neurons are generated from radial glial cells (RGCs), which exhibit two distinct modes of division during neurogenesis: (1) direct neurogenesis, where an RGC directly produces a neuron, and (2) indirect neurogenesis, where an RGC generates an intermediate neural progenitor (INP) that subsequently produces neurons. (B) Indirect neurogenesis is particularly prevalent during the early stages of neurogenesis. Neurons generated through both direct and indirect neurogenesis differentiate into similar neuronal types in a temporally ordered fashion. Indirect neurogenesis likely functions to amplify the production of similar types of neurons efficiently.
L6 CT, layer 6 corticothalamic projection neuron; SC, subcortical projection neuron; IT, intratelencephalic projection neuron.
A new Director-General has joined NIG as of December 1, 2024.
Dr. Shigeru Kondo came into office as the 11th Director-General of the National Institute of Genetics.
Under the new administration led by Dr. Kondo, the National Institute of Genetics aims to contribute to the further development of life science studies.

Xiaoxuan Zhu and Yuki Hatoyama received the “Excellent Poster Award”
Dr. Xiaoxuan Zhu (Postdoctoral Researcher) and Mr. Yuki Hatoyama (5th-year PhD student at SOKENDAI and JSPS Research Fellow DC2) from the Laboratory of Molecular Cell Engineering (Kanemaki Lab) presented posters at the 12th 3R+3C International Symposium held in Fukuoka from November 18th to 22nd, 2024. Both were honored with the “Excellent Poster Award.”
The 3R+3C International Symposium is a biennial international conference focused on research in DNA Replication, Repair, and Recombination (3R), as well as Chromatin, Chromosomes, and the Cell Cycle (3C).

Left, Zhu-san; Right, Hatoyama-san
Katsuhiko Minami and Masa A. Shimazoe won the “Excellent Poster Award”
Katsuhiko Minami (SOKENDAI D5 student and JSPS Special Researcher) and Masa A. Shimazoe (SOKENDAI D3 student and JSPS Special Researcher) from Genome Dynamics Laboratory received the “Excellent Poster Award” at the 12th 3R+3C International Symposium held in Fukuoka, Japan, on November 18th – 22nd.
The 3R+3C International Symposium is an international research meeting on 3R+3C, which stands for DNA replication, repair, recombination plus chromatin, chromosome, and cell cycle.

Left, Minami-san; Right, Shimazoe-san
Character displacement or priority effects: immigration timing can affect community assembly with rapid evolution
Character displacement or priority effects: immigration timing can affect community assembly with rapid evolution
Keiichi Morita, Masato Yamamichi
Proceedings of the Royal Society B: Biological Sciences (2024) 291: 20242145. DOI:10.1098/rspb.2024.214
![]() Press release (In Japanese only)
Press release (In Japanese only)
Understanding how biological communities assemble in the presence of rapid evolution is becoming an important topic in ecology. Previous studies demonstrated that community assembly can be affected by two types of eco-evolutionary dynamics: evolution-mediated priority effect (EPE) and ecological character displacement (ECD). In EPE, early-arriving species prevent colonization of late-arriving species via local adaptation (i.e. community monopolization), whereas ECD promotes species coexistence by niche partitioning. Researchers tended to discuss the two processes separately, but it should be possible for those processes to operate in the same system depending on various conditions. Here, we developed a theoretical framework that integrates the two processes by using a simple two-species competition model with eco-evolutionary feedback. We revealed that, when an early-arriving species evolves, the difference in immigration timing between the early-arriving and a late-arriving species can be a key parameter. When the difference is small, ECD occurs because insufficient local adaptation of the early-arriving species allows colonization of the late-arriving species. When the difference is large, however, EPE occurs because niche pre-emption by local adaptation of the early-arriving species prevents colonization of the late-arriving species. Further theoretical and empirical studies will be important to better understand eco-evolutionary community assembly with ECD and EPE.
Summer Internship at NIG “NIG-INTERN2025”
A complete classification of evolutionary games with environmental feedback
A complete classification of evolutionary games with environmental feedback
Hiromu Ito & Masato Yamamichi.
PNAS Nexus (2024). 3 (11): pgae455. DOI:10.1093/pnasnexus/pgae455
![]() Press release (In Japanese only)
Press release (In Japanese only)
A tragedy of the commons, in which rational behavior of individuals to maximize their own payoffs depletes common resources, is one of the most important research topics in game theory. To better understand the social dilemma problem, recent studies have developed a theoretical framework of feedback-evolving game where individual behavior affects an environmental (renewable) resource and the environmental resource changes individual payoffs. While previous studies assumed that the frequency of defectors increases (prisoner’s dilemma [PD] game) when the environmental resource is abundant to investigate an oscillating tragedy of the commons, it is also possible for other types of game to produce the social dilemma. In this paper, we extend the feedback-evolving game by considering not only PD game, but also the other three game structures when the environmental resource is replete for a reasonably complete classification. The three games are Chicken game where defectors and cooperators coexist through minority advantage, Stag-Hunt (SH) game with minority disadvantage, and Trivial game where the frequency of cooperators increases. In addition, we utilize a dilemma phase plane to visually track (transient) dynamics of game structure changes. We found that an emergent initial condition dependence (i.e. bistability) is pervasive in the feedback-evolving game when the three games are involved. We also showed that persistent oscillation dynamics arise even with Chicken or SH games in replete environments. Our generalized analysis will be an important step to further extend the theoretical framework of feedback-evolving game to various game situations with environmental feedback.
【For Graduate Students Only】 Guidelines for NIG Special Collaborative Research Students 2025
Making mirror-symmetric organ
Sawa Group / Multicellular Organization Laboratory
Distinct functions of three Wnt proteins control mirror-symmetric organogenesis in the C. elegans gonad
Shuhei So, Masayo Asakawa and Hitoshi Sawa
eLife (2024) Nov 1:13:e103035 DOI:10.7554/eLife.103035
Organogenesis requires the proper production of diverse cell types and their positioning/migration. However, the coordination of these processes during development remains poorly understood. The gonad in C. elegans exhibits a mirror-symmetric structure guided by the migration of distal tip cells (DTCs), which result from asymmetric divisions of somatic gonadal precursors (SGPs; Z1 and Z4). We found that the polarity of Z1 and Z4, which possess mirror-symmetric orientation, is controlled by the redundant functions of the LIN-17/Frizzled receptor and three Wnt proteins (CWN-1, CWN-2, and EGL-20) with distinct functions. In lin-17 mutants, CWN-2 promotes normal polarity in both Z1 and Z4, while CWN-1 promotes reverse and normal polarity in Z1 and Z4, respectively. In contrast, EGL-20 inhibits the polarization of both Z1 and Z4. In lin-17 egl-20 cwn-2 triple mutants with a polarity reversal of Z1, DTCs from Z1 frequently miss-migrate to the posterior side. Our further analysis demonstrates that the mis-positioning of DTCs in the gonad due to the polarity reversal of Z1 leads to mis-migration. Similar mis-migration was also observed in cki-1(RNAi) animals producing ectopic DTCs. These results highlight the role of Wnt signaling in coordinating the production and migration of DTCs to establish a mirror-symmetric organ.
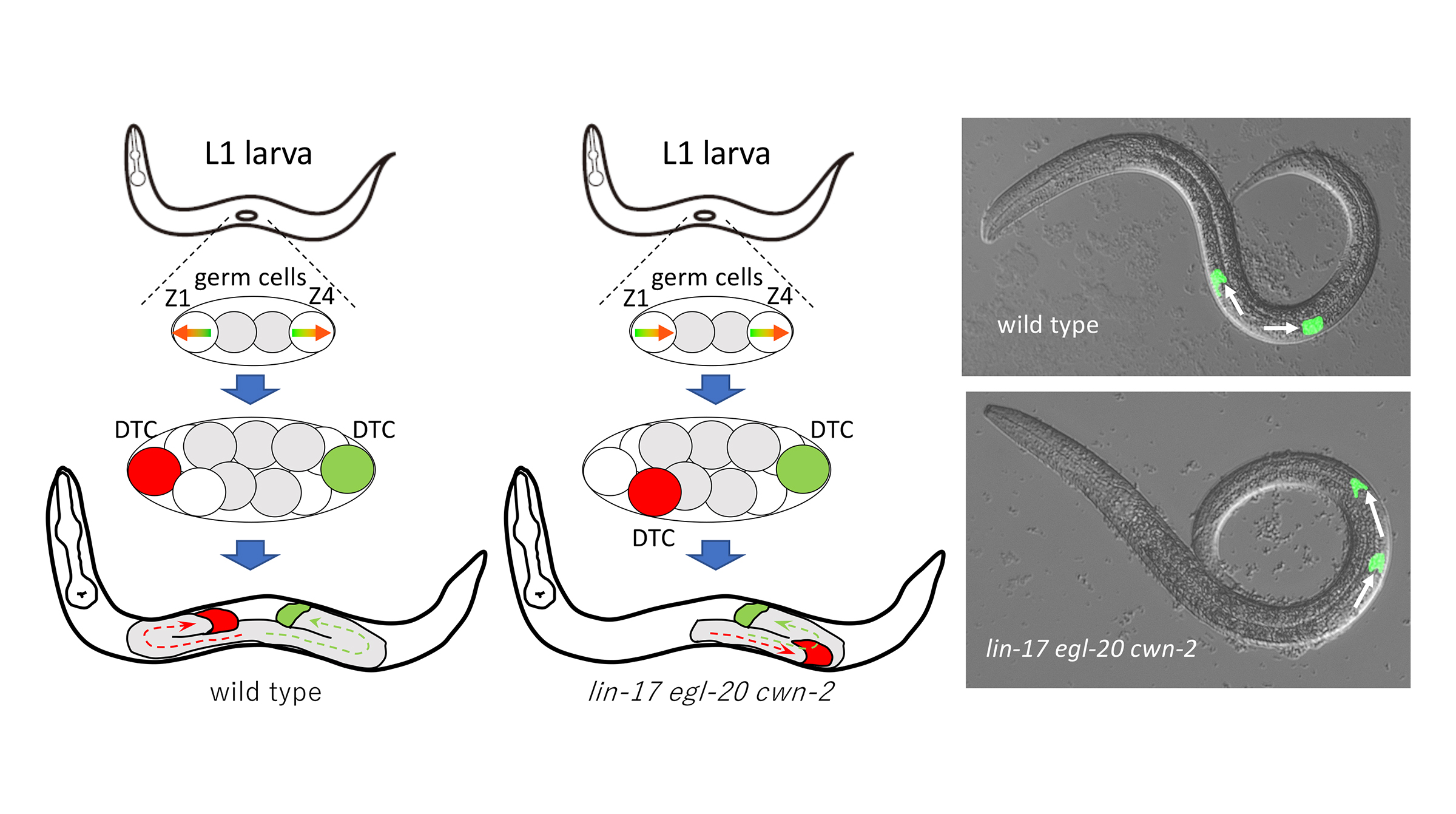
Figure: Left: Development of wild type gonad in C. elegans. Z1 and Z4 cells have mirror-symmetric polarity and produces two DTCs at the distal ends of the gonad. Each DTC migrate distally. Center: In lin-17 egl-20 cwn-2 mutants, Z1 polarity is reversed, producing DTC at the center of the gonad which migrate posteriorly. Right:Images of migrating DTCs (green).
Dr. Ken-ichi NONOMURA promoted to professor
Associate professor Ken-ichi NONOMURA (Plant Cytogenetics Laboratory) was promoted to professor as of November 1, 2024. Congratulations!
> NONOMURA, Ken-ichi : Plant Cytogenetics Laboratory, Department of Gene Function and Phenomics

NONOMURA, Ken-ichi Professor
Cataloging the availability of whole genome sequence information for endangered species in Japan: where is the cold spot?
Kuraku Group / Molecular Life History Laboratory
Genome assembly catalog for species in the Japanese Red List: unlocking endangered biodiversity through genomic inventory
Kryukov, K., Nakahama, N., and Kuraku, S.
F1000Research (2024) 13, 583 DOI:10.12688/f1000research.149793.2

Genomic information on another subspecies of the same species, Vespa crabro crabro, was recently obtained by a British institute and shown to be consistent with the previously reported karyotype (2n=50).
Whole genome sequence reading, which can be regarded as a catalog of DNA information, is a prerequisite for an accurate understanding of the genetic diversity that is key to the survival of a species; DNA sequences also contain information on the molecular basis that determines the ecological, morphological, and behavioral characteristics of a species. In both ways, whole genome sequencing is essential for understanding biodiversity, yet this information is available for only a handful of species. Dr. Kirill Kryukov of Joint Support-Center for Data Science Research of Research Organization of Information and Systems (ROIS-DS), Dr. Naoyuki Nakahama of the University of Hyogo and Museum of Nature and Human Activities, Hyogo, and Dr. Shigehiro Kuraku of the National Institute of Genetics have therefore focused their attention on rare species among the wild organisms living in Japan, and have developed a database, Genome sequence data availability for the Japanese Red List, which lists the accumulation status of whole genome sequence information.
This database catalogs the registration status of NCBI whole genome sequence information for rare species based on the “Red List 2020 of the Ministry of the Environment (In Japanese only)” and the “Red List of Marine Organisms of the Ministry of the Environment”, and the summarized information is made freely accessible online. The information can be viewed as a table in a web browser. The status of accumulation can be checked at a glance not only by species or subspecies, but also by taxonomic units such as “mammals,” “insects,” and “fungi” (Fig. 1). The system is also equipped with a function to regularly update the display content to reflect new information registered daily from all over the world. In addition, by downloading the files in TSV format, it can be used for secondary data analysis.

Figure1: Status of genome sequence data availability by taxon
One of the tables from the the top page of the database. The rightmost column shows the percentage of species for which genome information has been developed among the species considered to be rare in Japan. The percentage is generally very low, although it varies from taxon to taxon.
Whole genome sequencing requires not only suitable samples for extracting high molecular weight DNA and capturing the nuclear 3D structure of DNA molecules, but also the technical capability to develop sequence information that can withstand extensive use. Although numerous technological innovations and optimizations have reduced the cost and time required for calculations, the cost sometimes runs into the millions of yen, depending on the total number of bases in the genome, and also requires the labor of specialized technical personnel. Despite these limitations, it is important to steadily accumulate information, and there are many cases overseas where national governments are taking the initiative in organizing the priorities of indigenous species and preemptively acquiring genome information. The product of this study are expected to serve as a guide for Japan to vigorously promote such efforts by identifying priorities based on the recognition of the bias in the sequence data availability, and to efficiently acquire information in a more preemptive manner.
Establishment of mouse lines useful for endogenous protein degradation.
Saga Group / Mammalian Development Laboratory
Kanemaki Group / Molecular Cell Engineering Laboratory
Establishment and characterization of mouse lines useful for endogenous protein degradation via an improved auxin-inducible degron system (AID2).
Makino-Itou H, Yamatani N, Okubo A, Kiso M, Ajima R, Kanemaki MT, Saga Y.
Development, Growth and Differentiation (2024) Sep;66(7):384-393. DOI:10.1111/dgd.12942
The development of new technologies opens new avenues in the research field. Currently, conditional gene knockout strategies are employed to examine temporal and spatial gene function. However, phenotypes are sometimes not observed because of the time required for depletion due to the long half-life of the target proteins. Protein knockdown using an improved auxin-inducible degron system, AID2, overcomes such difficulties owing to rapid and efficient target depletion. Here, we established several mouse lines useful for AID2-medicated protein knockdown, which include knock-in mouse lines in the ROSA26 locus; one expresses TIR1(F74G), and the other is the reporter expressing AID-mCherry. We also established a germ-cell-specific TIR1 line and confirmed the protein knockdown specificity. In addition, we introduced an AID tag to an endogenous protein, DCP2 via the CAS9-mediated gene editing method. We confirmed that the protein was effectively eliminated by TIR1(F74G), which resulted in the similar phenotype observed in knockout mouse within 20 h.
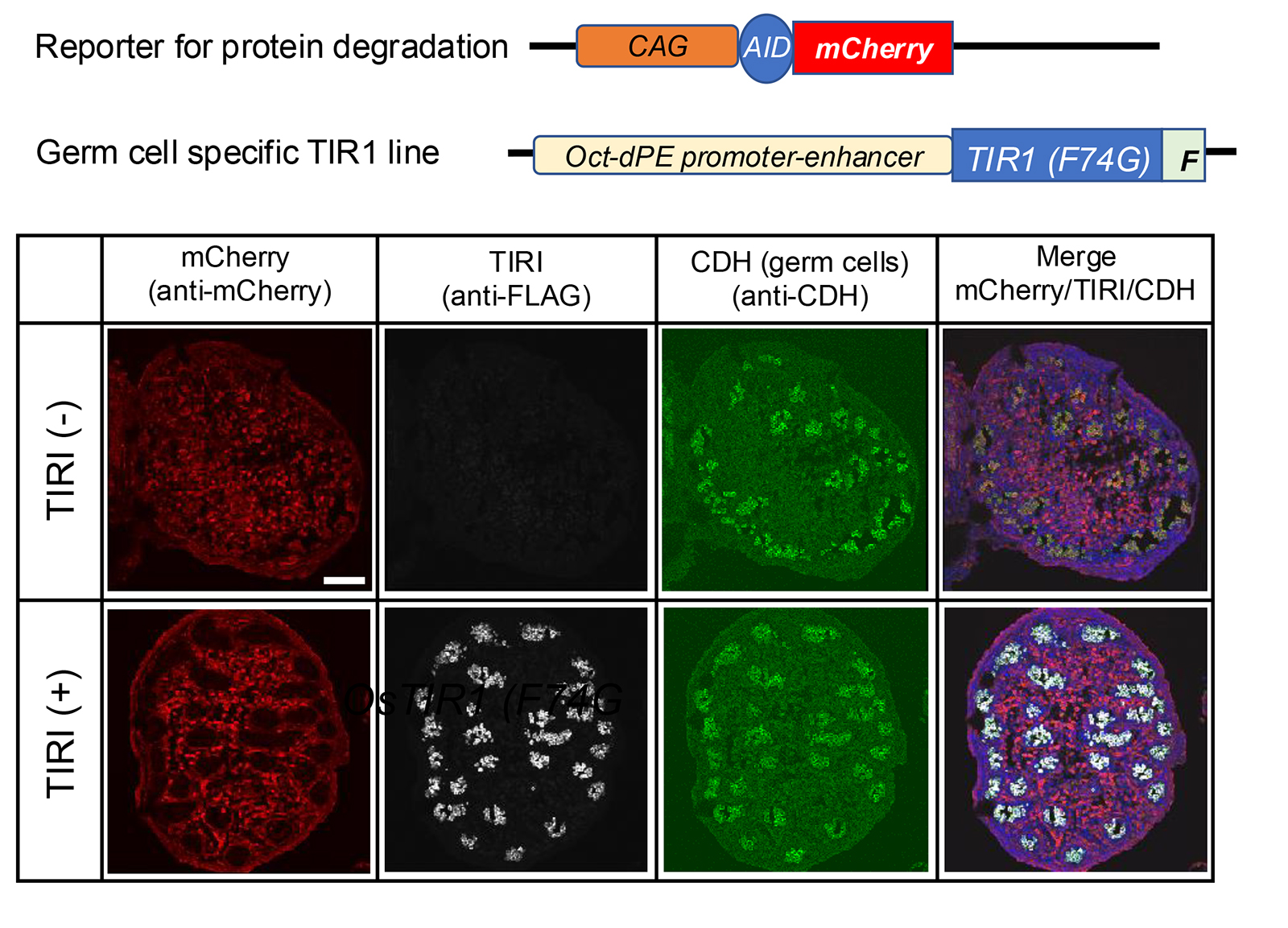
Figure: The germ-cell-specific TIR1 line was crossed with the reporter line TG-CAG-AID-mCherry. Immunofluorescence images of embryonic testes (E15.5) prepared from a pregnant CAG-AID-mCherrymother crossed with an Oct-dPE-TIR1(F74G)-FLAG male. TIR1(-) means sample without Oct-dPE-TIR1(F74G) and TIR1(+) means double transgenic sample. The mother was subjected to 5-Ph-IAA injection at E14.5. Antibodies used were anti-mCherry, anti-FLAG, and anti-E-cadherin (CDH). The scale bar indicates 100 mm.
Appointment of the New Director-General of the National Institute of Genetics
October 24, 2024
Upon the expiration of the term of the present Director-General of the National Institute of Genetics (NIG), Research Organization of Information and Systems (ROIS) has decided to appoint Dr. Shigeru Kondo as the next Director-General of the NIG, through the deliberation of the Education and Research Council.
Dr. Kondo will arrive at his post on the 1st of December, 2024.
Term of Office: From the 1st of December, 2024 to the 30th of November, 2028 (a total of 4 years)
“Dean’s Award” awarded to Genetics Program student Bhim Bahadur Biswa
Mr. Bhim Bahadur Biswa, who graduated from SOKENDAI in September 2024 as a member of the Mouse Genomics Resource Laboratory, has been awarded the School of Life Science “Dean’s Award” for the first semester of 2024.
The Dean’s Award recognizes degree recipients who have conducted research worthy of commendation and reported their accomplishments in an outstanding doctoral thesis. The award was presented on September 27, 2024 during the graduation ceremony. In addition to the Dean’s Award, Mr. Biswa has also been awarded the Genetics Program’s Morishima Award.
・Thesis title : Role of gut bacteria in domestication of mice
Mr. Biswa has provided the following statement regarding the award.
“I am deeply honored to receive the Dean’s Award from the School of Life Sciences, SOKENDAI University, for my PhD research. This recognition underscores the significance of our work in understanding the role of the gut microbiome in the animal domestication process. I am immensely grateful for the guidance and support from my supervisors and lab members, as well as the collaborative environment fostered by the National Institute of Genetics, which has been crucial in advancing this project. This award motivates me to continue pursuing innovative research with the potential to make a lasting impact in the scientific community. “

Mr. Biswa, Bhim Bahadur (right) & Associate Professor Koide
Swing the cell to know the physics inside
Press release
Live-cell imaging under centrifugation characterized the cellular force for nuclear centration in the Caenorhabditis elegans embryo.
Makoto Goda, Michael Shribak, Zenki Ikeda, Naobumi Okada, Tomomi Tani, Gohta Goshima, Rudolf Oldenbourg, Akatsuki Kimura
Proceedings of the National Academy of Sciences (PNAS) (2024) 121 (43), e2402759121 DOI:10.1073/pnas.2402759121
![]() Press release (In Japanese only)
Press release (In Japanese only)
Genomic DNA containing genetic information is stored in the cell nucleus. The nucleus is often located near the center of the cell. This means that there is a force inside the cell that moves and maintains the nucleus at the center. Measuring the amount of force is a challenging task. In this study, the researchers succeeded in measuring the force that maintains the cell nucleus in the center by applying centrifugal force to the cell using a special microscope called centrifuge polarizing microscope (CPM). CPM enables researchers to observe cells while rotating at a high speed. Researchers found that when a cell is rotated at a high speed, the cell nucleus is displaced from the center of the cell. The greater the centrifugal force, the greater the displacement of the nucleus from the center of the cell. Another special microscope, called an orientation-independent differential interference contrast (OI-DIC) microscope, revealed the mass density of the cell nucleus and thus enabled the researcher to calculate the centrifugal force acting on the nucleus. From this relationship between force and displacement, the researchers succeeded in quantifying the tiny force generated inside the cell to keep the nucleus centered. Cells are crowded with high concentrations of large molecules such as proteins. It has been a mystery how a large structure, such as the cell nucleus, can move inside crowded cells. This research unraveled part of this mystery using CPM and OI-DIC microscopes through the international collaboration.
The Hox patterning system underlying the pectoral fin formation was established before the divergence of ray-finned and lobe-finned fishes
Technical Section / Phenotype Research Center / Cell Architecture Laboratory
The functional roles of zebrafish HoxA– and HoxD-related clusters in the pectoral fin development
Mizuki Ishizaka, Akiteru Maeno, Hidemichi Nakazawa,, Renka Fujii, Sae Oikawa, Taisei Tani, Haruna Kanno, Rina Koita, and Akinori Kawamura
Scientific Reports (2024) 14, 23602 DOI:10.1038/s41598-024-74134-9
Although the pectoral fins of ray-finned fishes and the forelimbs of tetrapods differ in morphology and function, they share a homology with a common evolutionary origin. Comparing the developmental mechanisms of these structures is expected to provide insights into the evolution of vertebrate appendage formation. Analysis of knockout mice has shown that the deletion of HoxA and HoxD clusters, including Hox 9-13 genes results in significant shortening of the forelimbs. Similarly, deleting hox13 genes in HoxA and HoxD-related clusters of zebrafish also led to morphological abnormalities in the pectoral fins. However, the role of hox genes other than hox13 in pectoral fin formation remains unknown. In this study, we generated multiple zebrafish mutants with combinatorial deletions of HoxA and HoxD-related clusters and found that the pectoral fins in these mutants were significantly shortened, mirroring the abnormalities observed in the knockout mice. Furthermore, detailed analysis of the pectoral fins of surviving adult hox mutant zebrafish using micro-CT scans revealed morphological abnormalities in regions of the pectoral fins that are homologous to the forelimbs. These results further support the hypothesis that the formation of pectoral fins utilizing Hox genes in HoxA and HoxD clusters was established in common ancestors before the divergence of ray-finned and lobe-finned fishes.
This research was conducted by a group led by Associate Professor Akinori Kawamura of the Division of Life Science, Graduate School of Science and Engineering, Saitama University, with support from NIG-JOINT (38A2019, 7A2020, 66A2021, 18A2022, 31A2023).
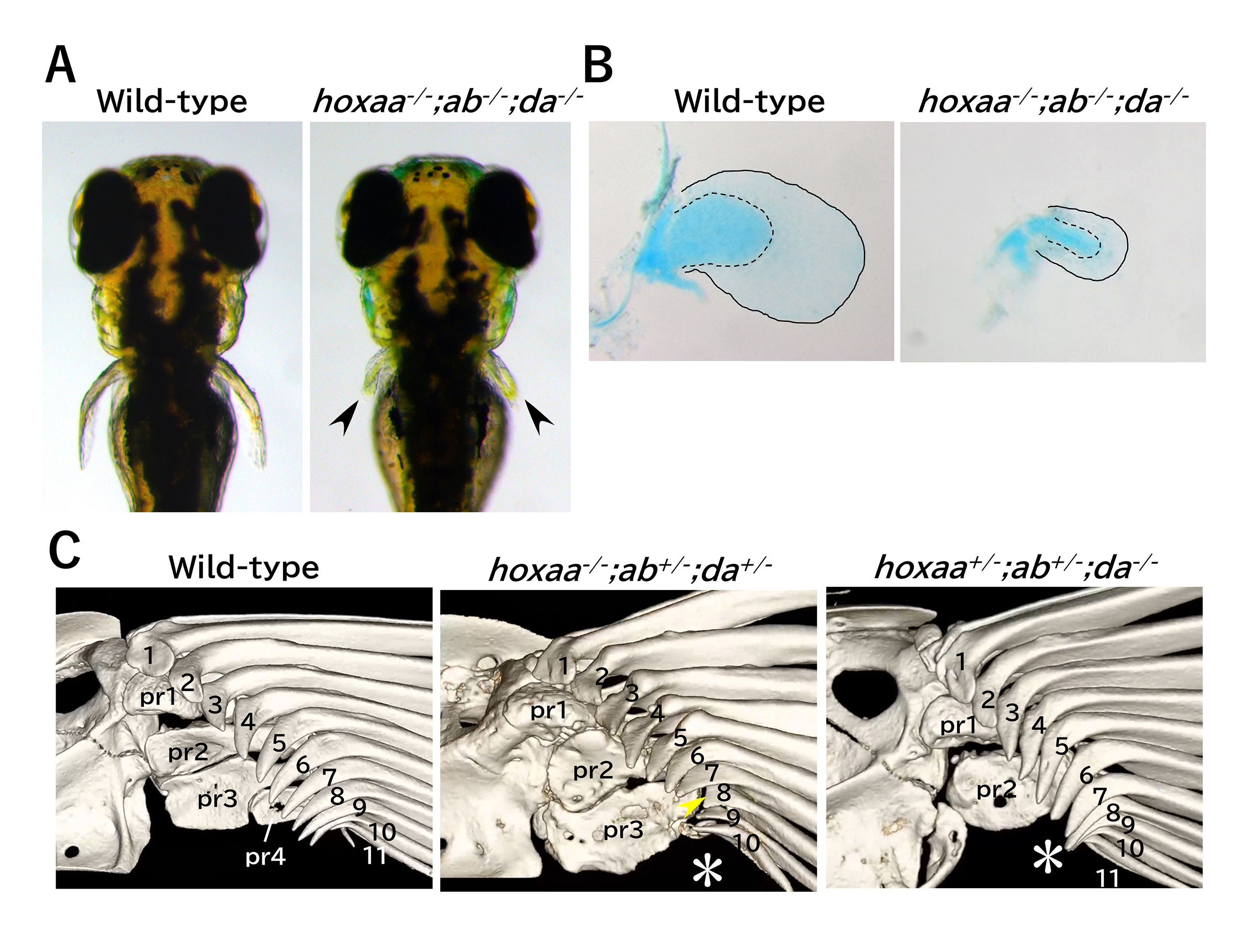
Figure: (A) Significant shortening of the pectoral fin in zebrafish hoxaa;ab;da cluster-delted mutants. 3-day post-fertilization. (B) Stained chondrocytes of the dissected pectoral fin (5-day). (C) Abnormal pectoral fin morphology in hox mutant adults revealed by micro-CT scan analysis.
Guideline for Application for 2025 NIG-JOINT(Joint Researchi and Research Meeting) *Application was closed
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Mettl1-dependent m7G tRNA modification is essential for maintaining spermatogenesis and fertility in Drosophila melanogaster
Press release
Mettl1-dependent m7G tRNA modification is essential for maintaining spermatogenesis and fertility in Drosophila melanogaster
Shunya Kaneko, Keita Miyoshi, Kotaro Tomuro, Makoto Terauchi, Ryoya Tanaka, Shu Kondo, Naoki Tani, Kei-Ichiro Ishiguro, Atsushi Toyoda, Azusa Kamikouchi, Hideki Noguchi, Shintaro Iwasaki, and Kuniaki Saito
Nature Communications (2024) 15, 8147 DOI:10.1038/s41467-024-52389-0
![]() Press release (In Japanese only)
Press release (In Japanese only)
Modification of guanosine to N7-methylguanosine (m7G) in the variable loop region of tRNA is catalyzed by the METTL1/WDR4 heterodimer and stabilizes target tRNA. Here, we reveal essential functions of Mettl1 in Drosophila fertility. Knockout of Mettl1 (Mettl1-KO) causes no major effect on the development of non-gonadal tissues, but abolishes the production of elongated spermatids and mature sperm, which is fully rescued by expression of a Mettl1-transgene, but not a catalytic-dead Mettl1 transgene. This demonstrates that Mettl1-dependent m7G is required for spermatogenesis. Mettl1-KO results in a loss of m7G modification on a subset of tRNAs and decreased tRNA abundance. Ribosome profiling shows that Mettl1-KO led to ribosomes stalling at codons decoded by tRNAs that were reduced in abundance. Mettl1-KO also significantly reduces the translation efficiency of genes involved in elongated spermatid formation and sperm stability. Germ cell-specific expression of Mettl1 rescues disrupted m7G tRNA modification and tRNA abundance in Mettl1-KO testes but not in non-gonadal tissues. Ribosome stalling is much less detectable in non-gonadal tissues than in Mettl1-KO testes. These findings reveal a developmental role for m7G tRNA modification and indicate that m7G modification-dependent tRNA abundance differs among tissues.
Mr. Biswa, Bhim Bahadur received the Morishima Award

On September 10, 2024, Mr. Biswa, Bhim Bahadur (Mouse Genomics Resource Laboratory, Koide Laboratory) received the Morishima Award from the Genetics Program, SOKENDAI.
The Morishima Award is given to students in the Genetics Program to honor their outstanding performances during PhD studies and to encourage further achievements.
・Biswa, Bhim Bahadur (Mouse Genomics Resource Laboratory, thesis advisor: Koide Tsuyoshi)
thesis title : Role of gut bacteria in domestication of mice















