Kimura Group • Cell Architecture Laboratory
Cell architectonics: how do the cells organize themselves?
Faculty
Research Summary
Cells are a beautiful example of architecture made by the nature. How such harmonious architecture is constructed ‘without an architect’ remains a mystery. This laboratory is studying, using approaches involving quantitative microscopy and theoretical modeling, the mechanisms underlying the movements of intracellular materials, such as organelle positioning and cytoplasmic streaming. We also investigate how neighboring cells physically interact to achieve appropriate arrangements. Through our studies, we aim to understand the secrets of constructing the cell.
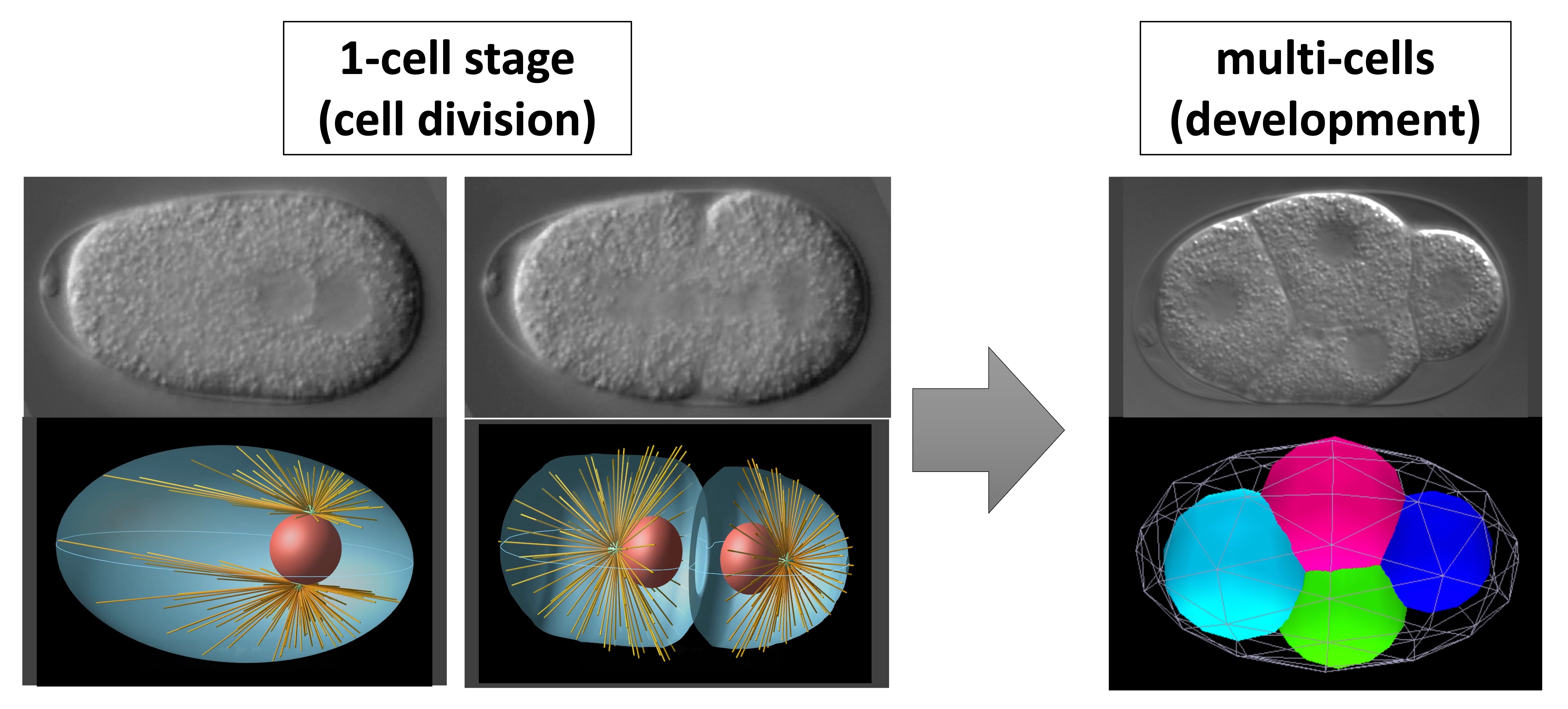
Selected Publications
Ishikawa T, Torisawa T, Wada H, Kimura A. Swirling instability mediated by elastic and hydrodynamic couplings in cytoplasmic streaming. PRX Life. 2025.3, 013008.
Goda M, Shribak M, Ikeda Z, Okada N, Tani T, Goshima G, Oldenbourg R, Kimura A. Live-cell imaging under centrifugation characterized the cellular force for nuclear centration in the Caenorhabditis elegans embryo. Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2402759121.
Kimura A. Quantitative Biology–A Practical Introduction. Springer 2022.

















