Maeshima Group • Genome Dynamics Laboratory
3D-organization and dynamics of human genome
Faculty
Research Summary
Our research interest is to know how a long string of human genome is three-dimensionally organized in the cell, and how the human genome is read out for cellular proliferation, differentiation and development. For this purpose, we are using a unique combination of molecular cell biology and biophysics, such as live-cell single molecule imaging, superresolution imaging, genomics and computational simulation.
Selected Publications
Hibino K, Sakai Y, Tamura S, Takagi M, Minami K, Natsume T, Shimazoe MA, Kanemaki MT, Imamoto N, Maeshima K. Single-nucleosome imaging unveils that condensins and nucleosome-nucleosome interactions differentially constrain chromatin to organize mitotic chromosomes. Nat Commun. 2024 Aug 21;15(1):7152.
Iida S, Ide S, Tamura S, Sasai M, Tani T, Goto T, Shribak M, Maeshima K. Orientation-independent-DIC imaging reveals that a transient rise in depletion attraction contributes to mitotic chromosome condensation. Proc Natl Acad Sci U S A. 2024 Sep 3;121(36):e2403153121.
Maeshima K, Iida S, Shimazoe MA, Tamura S, Ide S. Is euchromatin really open in the cell? Trends Cell Biol. 2024 Jan;34(1):7-17.
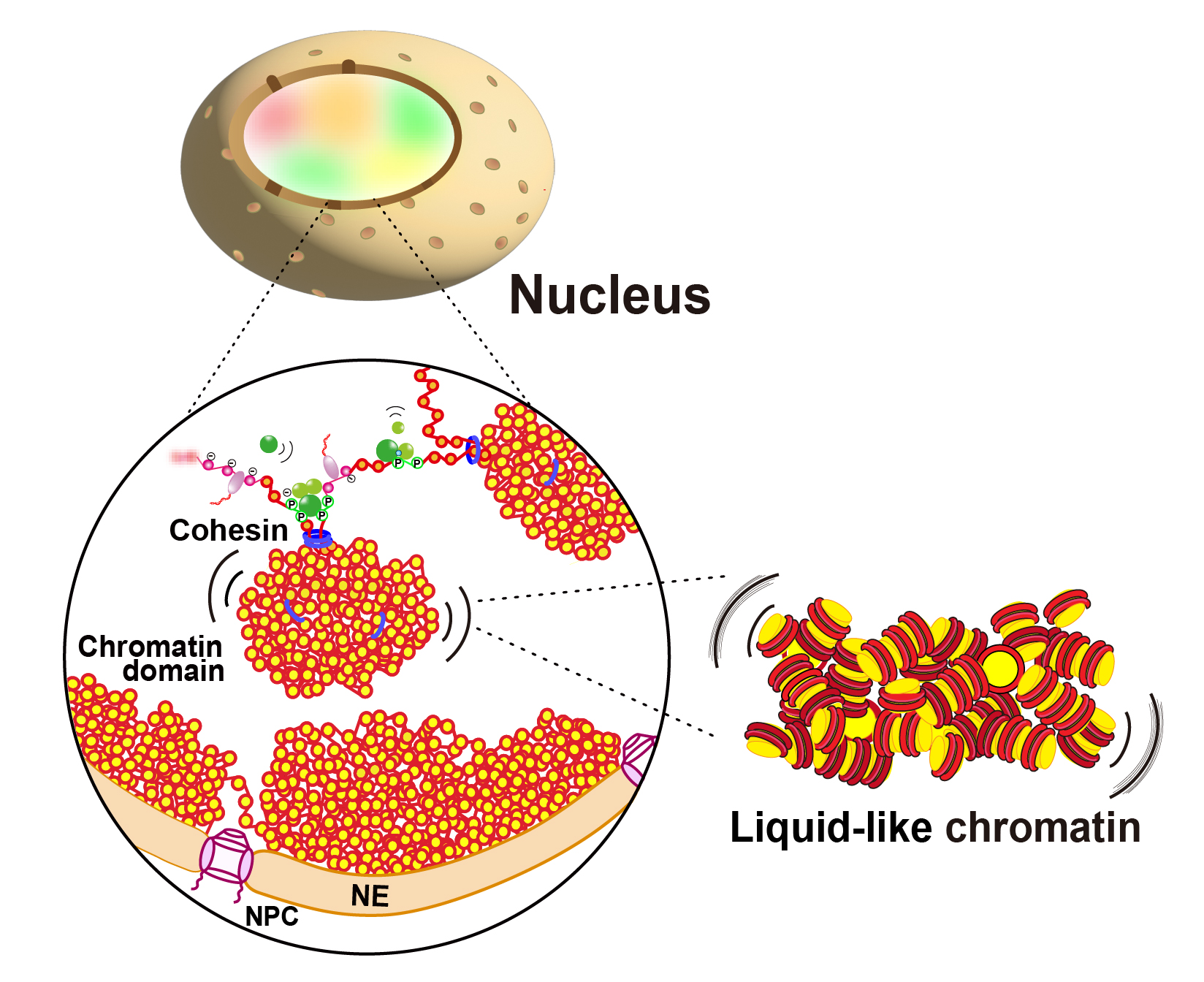
Nozaki T, Shinkai S, Ide S, Higashi K, Tamura S, Shimazoe MA, Nakagawa M, Suzuki Y, Okada Y, Sasai M, Onami S, Kurokawa K, Iida S, Maeshima K. Condensed but liquid-like domain organization of active chromatin regions in living human cells. Sci Adv. 2023 Apr 5;9(14):eadf1488.
















