Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Spinal RacGAP α-chimaerin is required to establish the midline barrier for proper corticospinal axon guidance
Press release
Acute inactivation of the replicative helicase in human cells triggers MCM8–9-dependent DNA synthesis
Toyoaki Natsume, Kohei Nishimura, Sheroy Minocherhomji, Rahul Bhowmick, Ian D. Hickson, Masato T. Kanemaki
Genes & Development DOI:10.1101/gad.297663.117
Pressrelease (In Japanese only)
Dr. Toyoaki Natsume and Prof. Masato Kanemaki at National Institute of Genetics, ROIS, together with the group led by Prof. Ian D. Hickson at University of Copenhagen, reported a new system to deal with failure in DNA replication. The finding was published in Genes & Development in advance of the print journal.
In cell proliferation, genomic DNA has to be precisely copied into two (DNA replication) before equal distribution to two daughter cells. For doing this, double-stranded DNA is unwound, the process of which is similar to open a zipper of your clothes (Figure 1A). Similar to pulling the ‘slider’ for opening a zipper, the replicative helicase known as MCM2–7 moves on DNA for unwinding double-stranded DNA. However, because human genomic DNA is very long (approx. 2 m per cell), it is challenging to entirely unwind the genomic DNA. Occasionally, the MCM2–7 helicase falls off from DNA when it encounters a roadblock (such as DNA damage). Because reloading of MCM2–7 is strictly inhibited during S phase (when cells carry out DNA replication), this might lead to incompletion of DNA replication, which causes the loss of genetic information from daughter cells unless the cells have a mechanism to deal with the problem.
In this study, the research groups observed how human cells responded to artificial removal of the MCM2–7 helicase by using the auxin-inducible degron (AID) technology that they had developed previously (the information about this technology is described here). They revealed that the MCM8–9 helicase, which is evolutionally related to the MCM2–7 helicase, promotes a non-canonical DNA synthesis as a backup system after removal of MCM2–7 (Figure 1B).
Many anticancer drugs kills cancer cells by inducing DNA lesions, which enhance removal of MCM2–7 from DNA during DNA replication. An inhibitor of the MCM8–9 helicase might enhance the effect of existing anticancer drugs by shutting off this backup system (Figure 2).
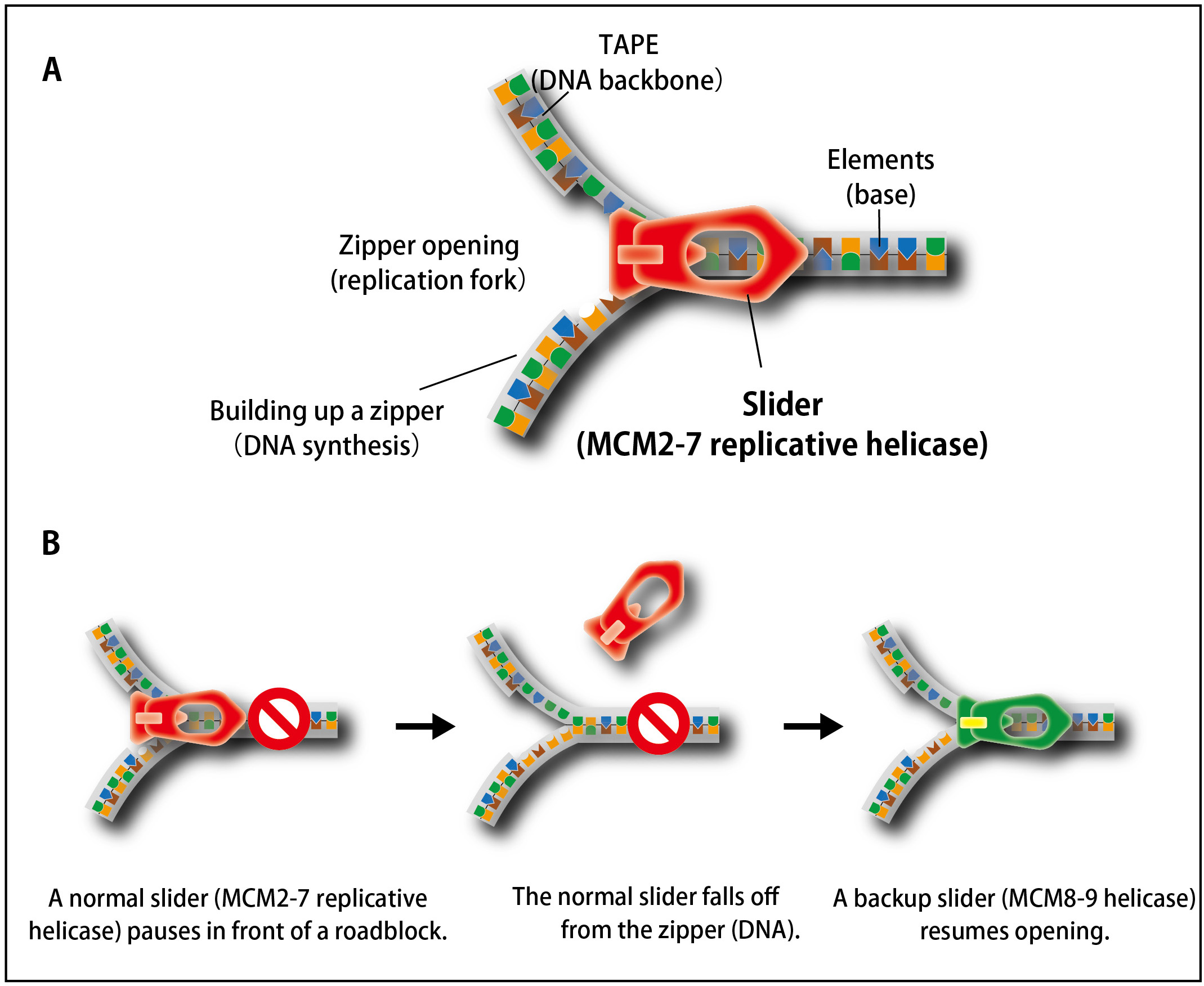
Figure 1. The process of unwinding double-stranded DNA during DNA replication is similar to that of opening a zipper of your clothes.
(A) Similar to the slider that opens a zipper, the MCM2–7 replicative helicase opens double-stranded DNA in cells.(B) When the replicative MCM2–7 helicase (a normal slider) encounters to a roadblock, it occasionally falls off from DNA. To continue DNA synthesis, cells recruit the MCM8–9 helicase as a backup slider.
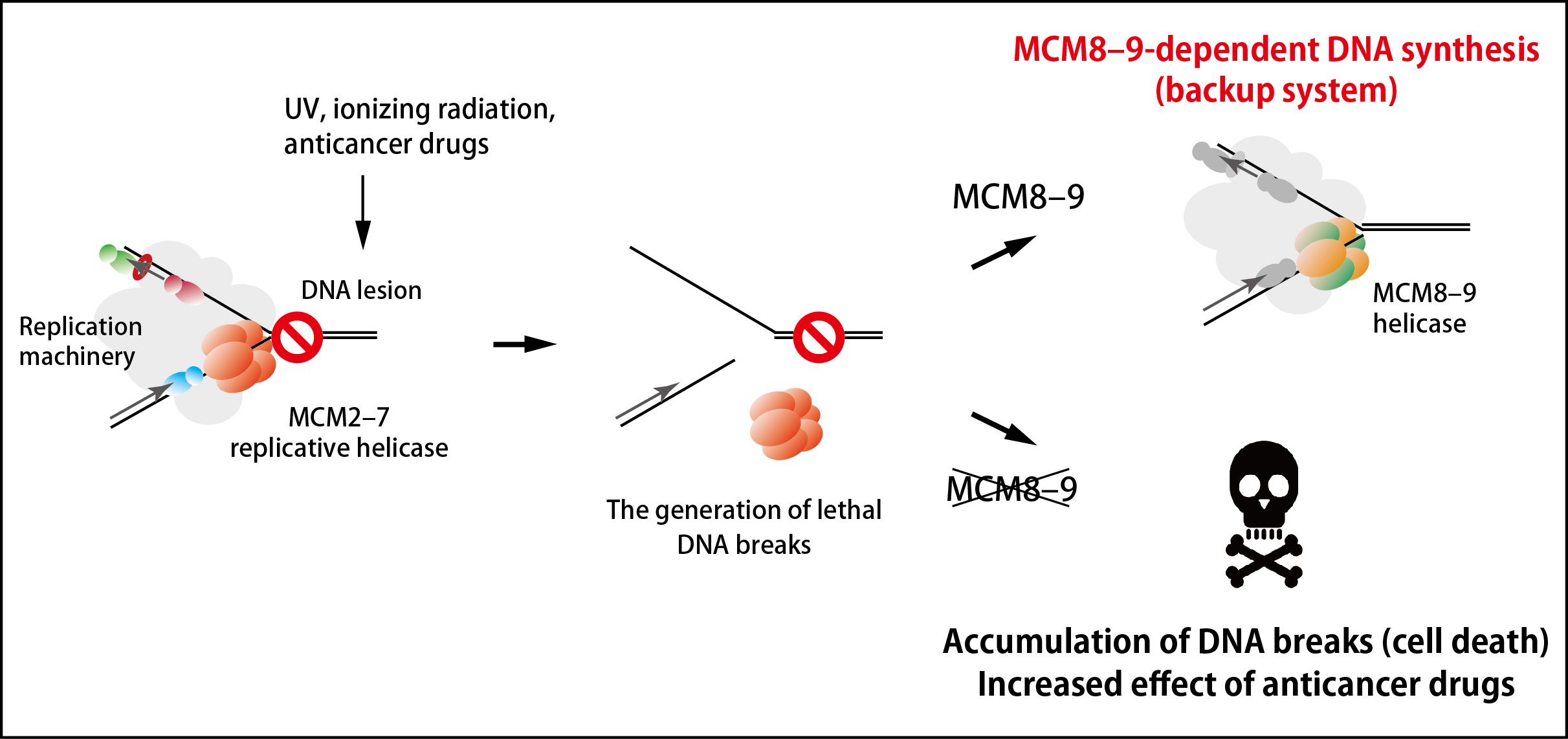
Figure 2. An MCM8–9-dependent backup system against failure in DNA replication.
The MCM2–7 replicative helicase falls off when encountered to a roadblock on DNA, leading to generation of DNA breaks. In this study, the research groups found that the MCM8–9 helicase continues DNA synthesis on behalf of the MCM2–7 (top right). If MCM8–9 does not work, the accumulation of DNA breaks results in cell death (bottom right).
Identification of a novel gene that increases cellulose synthesis in plants
Press release
A novel plasma membrane-anchored protein regulates xylem cell-wall deposition through microtubule-dependent lateral inhibition of Rho GTPase domains
Yuki Sugiyama, Mayumi Wakazaki, Kiminori Toyooka, Hiroo Fukuda, Yoshihisa Oda
Current Biology DOI:10.1016/j.cub.2017.06.059
Press release (In Japanese only)
Cellulose is an indispensable material in many applications for our life and industry. Cellulose is the main component of plant cell walls. However, the regulatory mechanism of cell wall synthesis is poorly understood. Our research group identified a novel gene named IQD13 that promotes cell wall synthesis in the model plant, Arabidopsis. We found that IQD13 products stabilizes microtubules that serve as a scaffold of cellulose synthesis. We also found that IQD13 increases area that are active for cell wall synthesis at the cell surface.
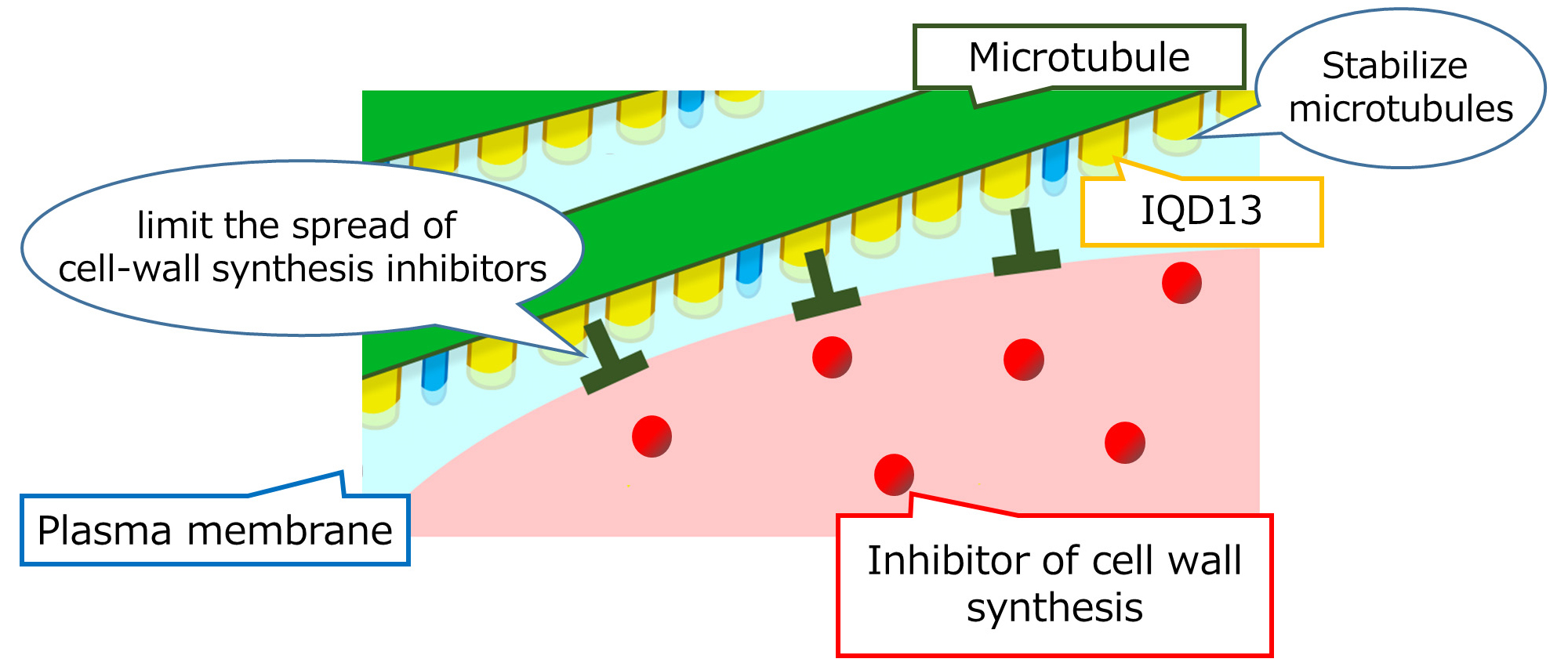
Figure: IQD13 prevents microtubule disruption and spread of the inhibitor of cell wall synthesis.
Spinal RacGAP α-chimaerin is required to establish the midline barrier for proper corticospinal axon guidance
Press release
Acute inactivation of the replicative helicase in human cells triggers MCM8–9-dependent DNA synthesis
Toyoaki Natsume, Kohei Nishimura, Sheroy Minocherhomji, Rahul Bhowmick, Ian D. Hickson, Masato T. Kanemaki
Genes & Development DOI:10.1101/gad.297663.117
Pressrelease (In Japanese only)
Dr. Toyoaki Natsume and Prof. Masato Kanemaki at National Institute of Genetics, ROIS, together with the group led by Prof. Ian D. Hickson at University of Copenhagen, reported a new system to deal with failure in DNA replication. The finding was published in Genes & Development in advance of the print journal.
In cell proliferation, genomic DNA has to be precisely copied into two (DNA replication) before equal distribution to two daughter cells. For doing this, double-stranded DNA is unwound, the process of which is similar to open a zipper of your clothes (Figure 1A). Similar to pulling the ‘slider’ for opening a zipper, the replicative helicase known as MCM2–7 moves on DNA for unwinding double-stranded DNA. However, because human genomic DNA is very long (approx. 2 m per cell), it is challenging to entirely unwind the genomic DNA. Occasionally, the MCM2–7 helicase falls off from DNA when it encounters a roadblock (such as DNA damage). Because reloading of MCM2–7 is strictly inhibited during S phase (when cells carry out DNA replication), this might lead to incompletion of DNA replication, which causes the loss of genetic information from daughter cells unless the cells have a mechanism to deal with the problem.
In this study, the research groups observed how human cells responded to artificial removal of the MCM2–7 helicase by using the auxin-inducible degron (AID) technology that they had developed previously (the information about this technology is described here). They revealed that the MCM8–9 helicase, which is evolutionally related to the MCM2–7 helicase, promotes a non-canonical DNA synthesis as a backup system after removal of MCM2–7 (Figure 1B).
Many anticancer drugs kills cancer cells by inducing DNA lesions, which enhance removal of MCM2–7 from DNA during DNA replication. An inhibitor of the MCM8–9 helicase might enhance the effect of existing anticancer drugs by shutting off this backup system (Figure 2).

Figure 1. The process of unwinding double-stranded DNA during DNA replication is similar to that of opening a zipper of your clothes.
(A) Similar to the slider that opens a zipper, the MCM2–7 replicative helicase opens double-stranded DNA in cells.(B) When the replicative MCM2–7 helicase (a normal slider) encounters to a roadblock, it occasionally falls off from DNA. To continue DNA synthesis, cells recruit the MCM8–9 helicase as a backup slider.

Figure 2. An MCM8–9-dependent backup system against failure in DNA replication.
The MCM2–7 replicative helicase falls off when encountered to a roadblock on DNA, leading to generation of DNA breaks. In this study, the research groups found that the MCM8–9 helicase continues DNA synthesis on behalf of the MCM2–7 (top right). If MCM8–9 does not work, the accumulation of DNA breaks results in cell death (bottom right).
Density imaging of chromatin in live cells
Biological Macromolecules Laboratory / Maeshima Group
Density imaging of heterochromatin in live cells using orientation-independent-DIC microscopy
Ryosuke Imai, Tadasu Nozaki, Tomomi Tani, Kazunari Kaizu, Kayo Hibino, Satoru Ide, Sachiko Tamura, Koichi Takahashi, Michael Shribak, and Kazuhiro Maeshima
Molecular Biology of the Cell, 2017 DOI:10.1091/mbc.E17-06-0359
In eukaryotic cells, highly condensed inactive/silenced chromatin has long been called “heterochromatin.” However, recent research suggests that such regions are in fact not fully transcriptionally silent, and that there exists only a moderate access barrier to heterochromatin. To further investigate this issue, it is critical to elucidate the physical properties of heterochromatin such as its total density in live cells. Here, using orientation-independent differential interference contrast (OI-DIC) microscopy, which is capable of mapping optical path differences, we investigated the density of the total materials in pericentric foci, a representative heterochromatin model, in live mouse NIH3T3 cells. We demonstrated that the total density of heterochromatin (208 mg/mL) was only 1.53-fold higher than that of the surrounding euchromatic regions (136 mg/mL) while the DNA density of heterochromatin was 5.5- to 7.5-fold higher. This surprisingly small difference may be due to that non-nucleosomal materials (proteins/RNAs)(∼120 mg/mL) are dominant in both chromatin regions. Monte Carlo simulation suggested that non-nucleosomal materials contribute to creating a moderate access barrier to heterochromatin, allowing minimal protein access to functional regions. Our OI-DIC imaging offers insight into the density of live cellular environments.
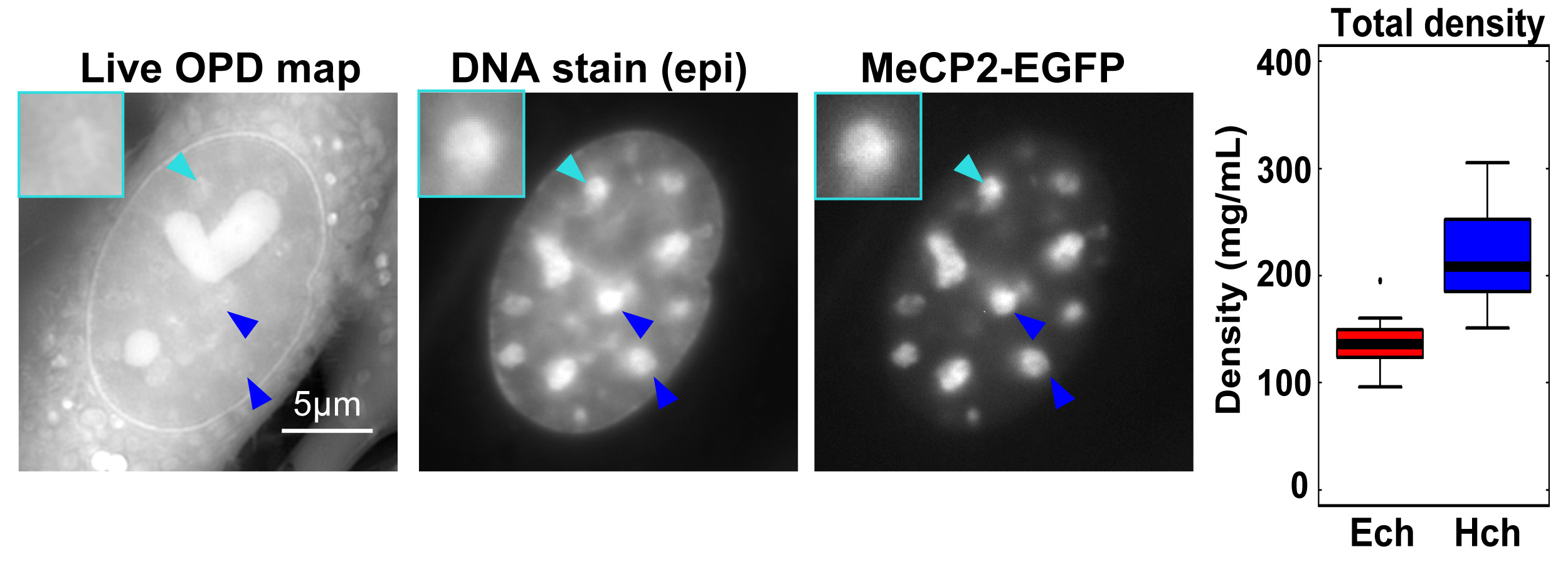
Typical images of the Optical Path Distance (OPD) map (reflecting density at each pixel) by OI-DIC, DNA staining, and MeCP2-EGFP (heterochromatin marker) signals in live NIH3T3 cells. Large foci seen in DNA staining and MeCP2-EGFP images (arrowheads) were assumed to be heterochromatin. Note that the OPD of the foci was similar or only slightly higher than that of the surrounding euchromatin. (Right) The analyzed total densities of pericentric heterochromatin foci (Hch, 208 mg/mL) and euchromatin (Ech, 136 mg/mL). The median density ratio between them was 1.53.
Summer Holiday (Aug. 14,15)
NIG will be closed from August 14 and August 15, 2017 for summer holiday.
Thank you for your understanding and cooperation.
Call for Collaboration Program at ROIS Joint Support-Center for Data Science Research (DS)
Call for Collaboration Program at ROIS Joint Support-Center for Data Science Research (DS)
Social hierarchy affects depression-like behavior and gene expression in mouse brain
Press release
Hierarchy in the home cage affects behaviour and gene expression in group-housed C57BL/6 male mice
Yasuyuki Horii, Tatsuhiro Nagasawa, Hiroyuki Sakakibara, Aki Takahashi, Akira Tanave, Yuki Matsumoto, Hiromichi Nagayama, Kazuto Yoshimi, Michiko T. Yasuda, Kayoko Shimoi, Tsuyoshi Koide
Scientific Reports Article number: 6991 (2017) DOI:10.1038/s41598-017-07233-5
Pressrelease (In Japanese only)
Social stress is one of the major causes of depression in humans. It is therefore important that the effects of social stress on behaviour and gene expression in the brain are studied. We performed experiments using male mice to analyse social hierarchy in groups of mice in cages and investigated emotional behaviour and hippocampal gene expression in the mice. We found significantly different emotional behaviour and expression of genes associated with the serotonergic system in dominant and subordinate mice. Treating the mice with a selective serotonin reuptake inhibitor antidepressant restored gene expression in subordinate mice and caused the emotional behaviour of the subordinate mice to be recovered, suggesting that alterations to the serotonergic system are key factors in phenotypic changes caused by the social ranks of subordinate mice. We believe that the results of this study are important because they improve our understanding of how social stress induces mental health problems in humans.
Spinal RacGAP α-chimaerin is required to establish the midline barrier for proper corticospinal axon guidance
Press release
Spinal RacGAP α-chimaerin is required to establish the midline barrier for proper corticospinal axon guidance
Shota Katori, Yukiko Noguchi-Katori, Shigeyoshi Itohara, Takuji Iwasato
Journal of Neuroscience 26 July 2017, 3123-16; DOI:10.1523/JNEUROSCI.3123-16.2017
Pressrelease (In Japanese only)
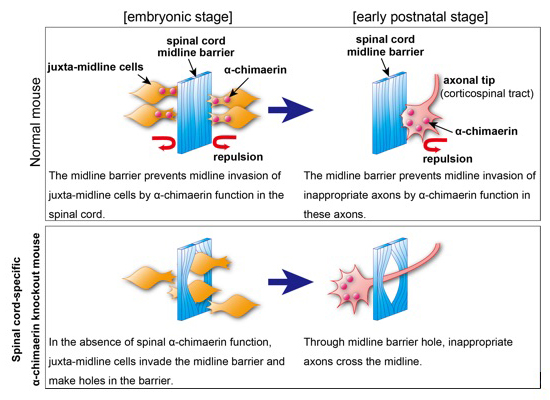
The midline barrier plays a critical role in midline axon guidance, which is fundamental to the formation of neural circuits that are responsible for proper left-right coordination of our body. Studies have revealed some of the mechanisms underlying how the midline barrier navigates axons. In contrast, the establishment of the midline barrier during embryonic development remains unclear. In this study, Katori et al. determined that α-chimaerin is required for the formation of an intact midline barrier. Spinal cord-specific α-chimaerin knockout mice had spinal midline barriers with numerous breaks (holes), through which corticospinal axons aberrantly crossed the midline. Katori et al. propose that α-chimaerin protects the midline barrier by mediating cell repulsive signaling in juxta-midline cells, which prevents these cells from invading the midline.
Source: Journal of Neuroscience 26 July 2017, 3123-16;
DOI:10.1523/JNEUROSCI.3123-16.2017
Systematic identification of regulatory elements derived from human endogenous retroviruses
Division of Human Genetics / Inoue Group
Systematic Identification and Characterization of Regulatory Elements Derived from Human Endogenous Retroviruses.
Jumpei Ito, Ryota Sugimoto, Hirofumi Nakaoka, Shiro Yamada, Tetsuaki Kimura, Takahide Hayano, and Ituro Inoue.
PLoS Genetics. Jul 12;13(7):e1006883. 2017. DOI:10.1371/journal.pgen.1006883
Human endogenous retroviruses (HERVs) have regulatory elements that possibly influence the transcription of host genes. We systematically identified these HERV regulatory elements (HERV-REs) based on publicly available datasets of ChIP-Seq. Overall, 794,972 HERV-REs were identified. Clustering analysis showed that HERVs can be grouped according to the TF binding patterns; HERV groups bounded by pluripotent TFs (e.g., SOX2, POU5F1, and NANOG), endoderm TFs (e.g., GATA4/6, SOX17, and FOXA1/2), hematopoietic TFs (e.g., SPI1, GATA1/2, and TAL1), and CTCF were identified. Regulatory elements of HERVs tended to locate nearby genes involved in immune responses, indicating that the regulatory elements play an important role in controlling the immune regulatory network. Finally, we constructed dbHERV-REs, an interactive database of HERV regulatory elements (http://herv-tfbs.com/). This study provides fundamental information in understanding the impact of HERVs on host transcription, and offers insights into the transcriptional modulation systems of HERVs.
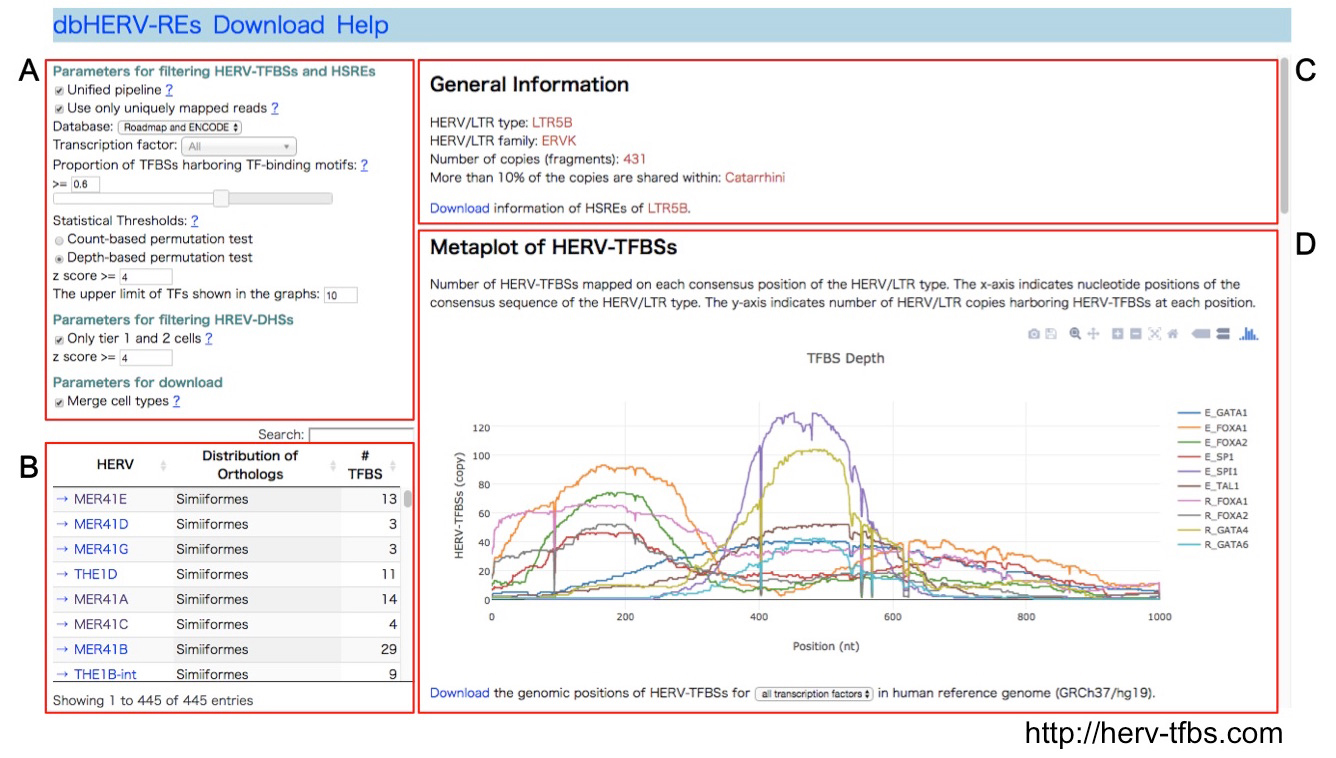
dbHERV-RE (http://herv-tfbs.com/). First, users choose a transcription factor and other parameters (A). Second, users select a type of HERVs (B). dbHERV-REs displays general information of the HERVs (phylogenetic classification, copy number, and insertion date) (C) and visualizes genetic positions of HERV-REs on the HERV sequence and the human reference genome (D).
Dynamic organization of chromatin domains in living cells.
Press release
Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging
Tadasu Nozaki, Ryosuke Imai, Mai Tanbo, Ryosuke Nagashima, Sachiko Tamura, Tomomi Tani, Yasumasa Joti, Masaru Tomita, Kayo Hibino, Masato T. Kanemaki, Kerstin S. Wendt, Yasushi Okada, Takeharu Nagai, and Kazuhiro Maeshima
Molecular Cell Published: July 13, 2017 DOI:10.1016/j.molcel.2017.06.018
Press release (In Japanese only)
The eukaryotic genome is organized within cells as chromatin. For proper information output, higher-order chromatin structures can be regulated dynamically. How such structures form and behave in various cellular processes remains unclear. Here, by combining super-resolution imaging (photoactivated localization microscopy [PALM]) and single-nucleosome tracking, we developed a nuclear imaging system to visualize the higher-order structures along with their dynamics in live mammalian cells. We demonstrated that nucleosomes form compact domains with a peak diameter of ∼160 nm and move coherently in live cells. The heterochromatin-rich regions showed more domains and less movement. With cell differentiation, the domains became more apparent, with reduced dynamics. Furthermore, various perturbation experiments indicated that they are organized by a combination of factors, including cohesin and nucleosome-nucleosome interactions. Notably, we observed the domains during mitosis, suggesting that they act as building blocks of chromosomes and may serve as information units throughout the cell cycle.
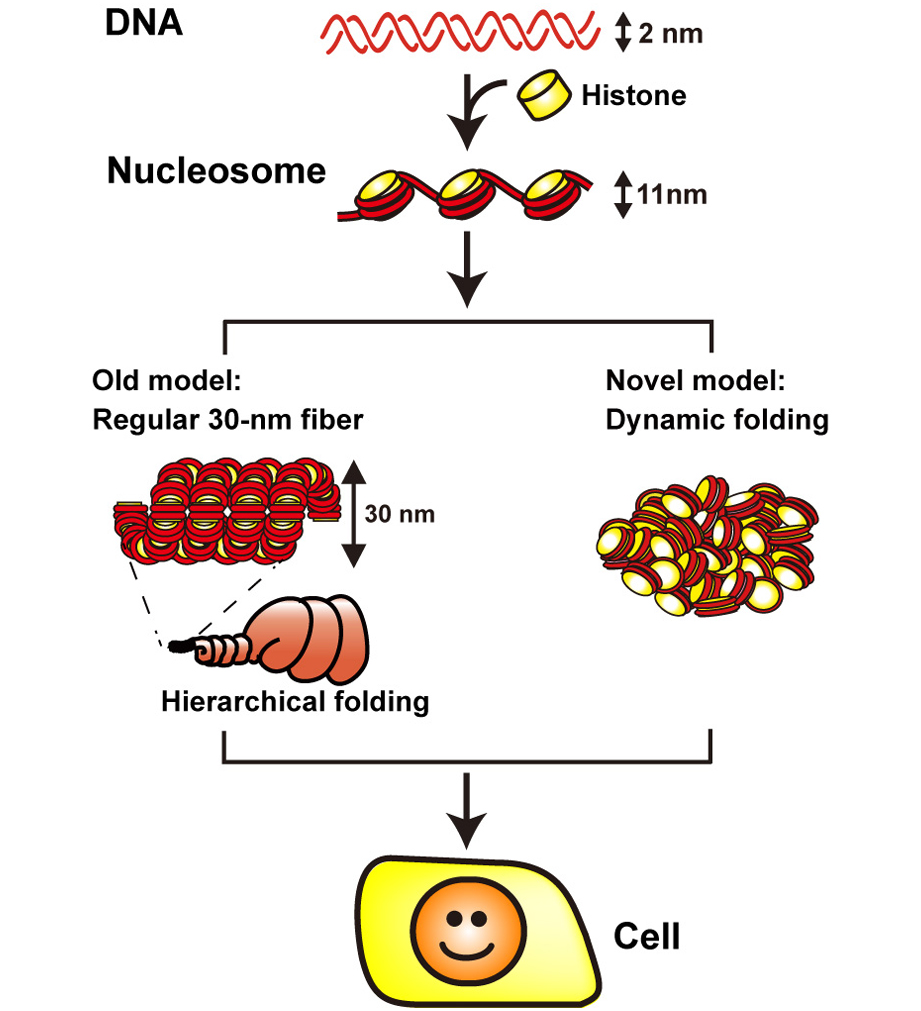
1. DNA (in red) is wrapped around core histone (in yellow) and forms a nucleosome structure (or 10-nm fiber). In the old model (left), the nucleosome fiber had long been assumed to fold into a 30-nm chromatin fiber, and subsequently into helically folded larger fibers (Hierarchical folding). In new current model (right), chromatin is composed of irregularly folded 10-nm fibers, without 30-nm chromatin fibers (dynamic folding) and stored in the cell.
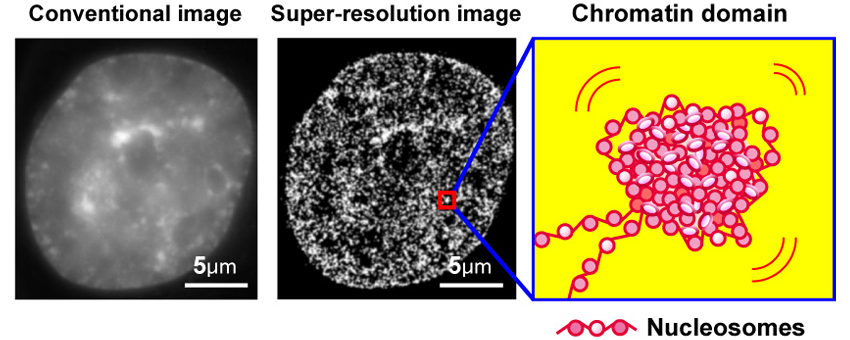
2. Left, conventional DNA staining image; center, super-resolution image of chromatin; a model of chromatin domain.
What makes domestic animals such as mice and dogs tame
Press release
Selective breeding and selection mapping using a novel wild-derived heterogeneous stock of mice revealed two closely-linked loci for tameness
Yuki Matsumoto, Tatsuhiko Goto, Jo Nishino, Hirofumi Nakaoka, Akira Tanave, Toshiyuki Takano-Shimizu, Richard F Mott, Tsuyoshi Koide
Scientific Reports 7, Article number: 4607 (2017) DOI:10.1038/s41598-017-04869-1
Pressrelease (In Japanese only)
Tameness play important role during the process of domestication, and can be divided into two potential components: motivation to approach humans (active tameness) and reluctance to avoid them (passive tameness). To understand the genetic basis associated with active tameness in mice we applied selective breeding of a genetically heterogeneous population that we founded by crossing eight wild mouse strains. As a result of selective breeding, the level of active tameness increased over the generations, compared to an unselected control experiment. We performed two selection and two control experiments. Genetic differences between the selected and control groups, measured using a high-dense array of single-nucleotide polymorphisms, were assessed using a computer simulation experiment. In one selection experiment we found a significant increase in the occurrence of a particular genomic segment present in just one of the founder strains, compared to the control groups. This selected region contains two loci related to active tameness and is syntenic to genomic regions which are known to be a region selected during dog domestication, suggesting that responsible genes in these loci are associated with active tameness in both mouse and dog.
A new laboratory established in the Center for Frontier Research
Yasuto MURAYAMA joined the Center for Frontier Science as of July 1, 2017.
MURAYAMA, Yasuto:Center for Frontier Research,Chromosome Biochemistry Laboratory

- MURAYAMA, Yasuto
Associate Professor
Center for Frontier Research is an incubation center to simultaneously develop two elements: human resources and new research fields. Promising young scientists conduct research as principal investigator (tenure-track associate professor) to explore new frontiers in genetics and related areas, taking advantage of NIG’s research infrastructure and various support systems.
Flexible DNA path in the MCM double hexamer loaded on DNA
Mammalian Development Laboratory / Saga Group
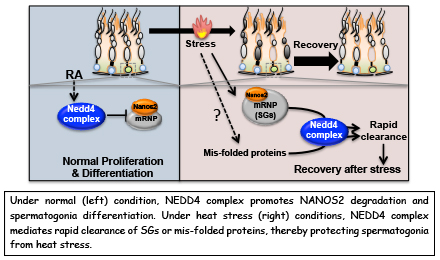
NEDD4 controls spermatogonial stem cell homeostasis and stress response by regulating messenger ribonucleoprotein complexes
Zhi Zhou, Hiroshi Kawabe, Atsushi Suzuki, Kaori Shinmyozu & Yumiko Saga
NATURE COMMUNICATIONS. 8, Article number: 15662 (2017) DOI:10.1038/ncomms15662
P bodies (PBs) and stress granules (SGs) are conserved cytoplasmic aggregates of cellular messenger ribonucleoprotein complexes (mRNPs) that are implicated in mRNA metabolism and play crucial roles in adult stem cell homeostasis and stress responses. However, the mechanisms underlying the dynamics of mRNP granules are poorly understood. Here, we report NEDD4, an E3 ubiquitin ligase, as a key regulator of mRNP dynamics that controls the size of the spermatogonial progenitor cell (SPC) pool. We find that NEDD4 targets an RNA-binding protein, NANOS2, in spermatogonia to destabilize it, leading to cell differentiation. In addition, NEDD4 is required for SG clearance. NEDD4 targets SGs and facilitates their rapid clearance through the endosomal–lysosomal pathway during the recovery period. Therefore, NEDD4 controls the turnover of mRNP components and inhibits pathological SG accumulation. Accordingly, we propose that a NEDD4-mediated mechanism regulates mRNP dynamics, and facilitates SPC homeostasis and viability under normal and stress conditions.This research was mainly conducted by Zhi Zhou who was a NIG postdoctoral fellow 2012-2014.
How do human cells repair a broken zipper of DNA? : A new backup system against failure in DNA replication
Press release
Acute inactivation of the replicative helicase in human cells triggers MCM8–9-dependent DNA synthesis
Toyoaki Natsume, Kohei Nishimura, Sheroy Minocherhomji, Rahul Bhowmick, Ian D. Hickson, Masato T. Kanemaki
Genes & Development DOI:10.1101/gad.297663.117
Pressrelease (In Japanese only)
Dr. Toyoaki Natsume and Prof. Masato Kanemaki at National Institute of Genetics, ROIS, together with the group led by Prof. Ian D. Hickson at University of Copenhagen, reported a new system to deal with failure in DNA replication. The finding was published in Genes & Development in advance of the print journal.
In cell proliferation, genomic DNA has to be precisely copied into two (DNA replication) before equal distribution to two daughter cells. For doing this, double-stranded DNA is unwound, the process of which is similar to open a zipper of your clothes (Figure 1A). Similar to pulling the ‘slider’ for opening a zipper, the replicative helicase known as MCM2–7 moves on DNA for unwinding double-stranded DNA. However, because human genomic DNA is very long (approx. 2 m per cell), it is challenging to entirely unwind the genomic DNA. Occasionally, the MCM2–7 helicase falls off from DNA when it encounters a roadblock (such as DNA damage). Because reloading of MCM2–7 is strictly inhibited during S phase (when cells carry out DNA replication), this might lead to incompletion of DNA replication, which causes the loss of genetic information from daughter cells unless the cells have a mechanism to deal with the problem.
In this study, the research groups observed how human cells responded to artificial removal of the MCM2–7 helicase by using the auxin-inducible degron (AID) technology that they had developed previously (the information about this technology is described here). They revealed that the MCM8–9 helicase, which is evolutionally related to the MCM2–7 helicase, promotes a non-canonical DNA synthesis as a backup system after removal of MCM2–7 (Figure 1B).
Many anticancer drugs kills cancer cells by inducing DNA lesions, which enhance removal of MCM2–7 from DNA during DNA replication. An inhibitor of the MCM8–9 helicase might enhance the effect of existing anticancer drugs by shutting off this backup system (Figure 2).

Figure 1. The process of unwinding double-stranded DNA during DNA replication is similar to that of opening a zipper of your clothes.
(A) Similar to the slider that opens a zipper, the MCM2–7 replicative helicase opens double-stranded DNA in cells.(B) When the replicative MCM2–7 helicase (a normal slider) encounters to a roadblock, it occasionally falls off from DNA. To continue DNA synthesis, cells recruit the MCM8–9 helicase as a backup slider.

Figure 2. An MCM8–9-dependent backup system against failure in DNA replication.
The MCM2–7 replicative helicase falls off when encountered to a roadblock on DNA, leading to generation of DNA breaks. In this study, the research groups found that the MCM8–9 helicase continues DNA synthesis on behalf of the MCM2–7 (top right). If MCM8–9 does not work, the accumulation of DNA breaks results in cell death (bottom right).
Flexible DNA path in the MCM double hexamer loaded on DNA
Division of Microbial Genetics / Araki Group
Flexible DNA Path in the MCM Double Hexamer Loaded on DNA
Kohji Hizume, Hiroaki Kominami, Kei Kobayashi, Hirofumi Yamada, and Hiroyuki Araki
Biochemistry. Publication Date (Web): May 1, 2017 DOI:10.1021/acs.biochem.6b00922
The formation of the pre-replicative complex (pre-RC) during the G1 phase, which is also called the licensing of DNA replication, is the initial and essential step of faithful DNA replication during the subsequent S phase. It is widely accepted that in the pre-RC, double-stranded DNA passes through the holes of two ring-shaped minichromosome maintenance (Mcm) 2–7 hexamers; however, the spatial organization of the DNA and proteins involved in the pre-RC formation is unclear.
A research group lead by Drs. Hiroyuki Araki and Kohji Hizume at NIG reconstituted pre-RC from purified DNA and proteins and visualized the complex using atomic force microscopy (AFM). Higher-resolution imaging of the pre-RC was performed by using FM-AFM in collaboration with the group of Professor Hirofumi Yamada at Kyoto University, and successfully detected two globules (two MCM hexamers) of the elliptical particle formed on DNA.
Analyses through AFM observation revealed that the DNA does not completely pass through both holes of the MCM hexamers, possibly because the DNA exited from the gap between Mcm2 and Mcm5. A DNA loop fastened by the MCM double hexamer was detected in pre-RC samples reconstituted from purified proteins as well as those purified from yeast cells. These results suggest that a higher-order architecture of the loaded MCM hexamers and DNA strands during licensing of DNA replication, which have not been proposed before.
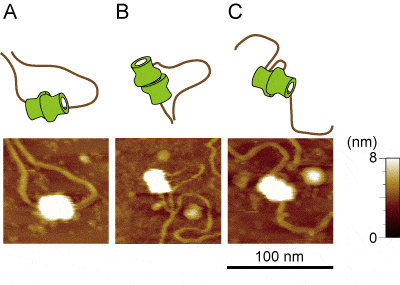
Loaded MCM–DNA complex visualized at higher resolution using FM-AFM. It was detected that the DNA passes through two MCM hexamers (A) as it is widely accepted. However, these were also detected that the DNA does not completely pass through both the MCM hexamers (B) and that DNA was fastened by the MCM double hexamer and form loop (C).
‘Eating with the eyes’ is hard-wired in the brain
Press release
Activation of the hypothalamic feeding centre upon visual prey detection
Akira Muto, Pradeep Lal, Deepak Ailani, Gembu Abe, Mari Itoh, Koichi Kawakami
Nature Communications 8, Article number: 15029 (2017) DOI:10.1038/ncomms15029
Pressrelease (In Japanese only)
Have you ever wondered why just seeing food can make your mouth start to water? By visualizing neuronal activity in specific areas of the zebrafish brain, scientists at the National Institute of Genetics (NIG) in Japan have revealed a direct link between visual perception of food and feeding motivation. The study, published in the April 20, 2017 issue of Nature Communications, suggests that “eating with the eyes” is deeply rooted in evolution.
This study was supported by JSPS KAKENHI Grant Numbers JP25290009 and JP25650120, and also partly supported by JSPS KAKENHI Grant Numbers JP15H02370 and JP16H01651, and NBRP from Japan Agency for Medical Research and Development (AMED). This work was also supported in part by the Center for the Promotion of Integrated Sciences (CPIS) of SOKENDAI.
Click here for article
https://www.eurekalert.org/pub_releases/2017-04/rooi-wt041717.php
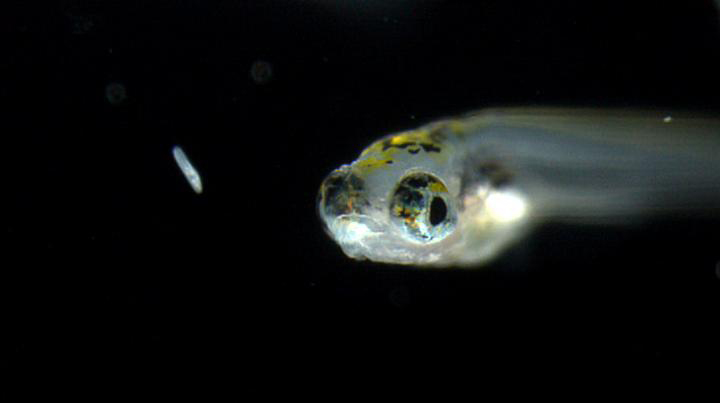
THIS IS A ZEBRAFISH LARVA TRYING TO CATCH PREY.
▶This study is based on the previous study.
Regulation of internode patterning and vein anastomosis in maize stems.
Experimental Farm / Nonomura Group
Mammalian Genetics Laboratory / Shiroishi Group
KNOTTED1 Cofactors, BLH12 and BLH14, Regulate Internode Patterning and Vein Anastomosis in Maize
Katsutoshi Tsuda, Maria-Jazmin Abraham-Juarez, Akiteru Maeno, Zhaobin Dong, Dale Aromdee, Robert Meeley, Toshihiko Shiroishi, Ken-ichi Nonomura and Sarah Hake.
The Plant Cell. published online April 5, 2017 DOI:10.1105/tpc.16.00967
Monocot stems lack the vascular cambium and instead have characteristic structures in which intercalary meristems generate internodes and veins remain separate and scattered. Developmental processes of these unique structures, however, have been poorly described. We found that maize BELL1-like homeodomain transcription factors, BLH12 and BLH14, have redundant but important roles in stem development. BLH12/14 interact with the shoot meristem regulator, KNOTTED1 (KN1) in vivo, and accumulate in overlapping domains in shoot meristems, young stems and provascular bundles. In addition to defects in the maintenance and development of various shoot meristems, blh12/14 double mutant showed unique abnormalities in the stem including shortened internodes and the reduced vein number. Detailed observation of BLH12/14 accumulation patterns and of stem inner structures using micro-computed tomography (CT) suggested that BLH12/14 (1) maintain intercalary meristems at the bottom of internodes to provide internodal cells as differentiated progenies, and (2) prevent precocious anastomosis of provascular bundles in young stems to ensure the production of sufficient independent veins. This work is the collaborative work between Katsutoshi Tsuda in Experimental Farm and Prof. Sarah Hake in University of California, Berkeley, and was supported by JSPS KAKENHI JP16K18637. Micro-CT scanning performed by Akiteru Maeno in Mammalian Genetics Lab enabled to capture the detailed view of inner stem structures and vein networks.
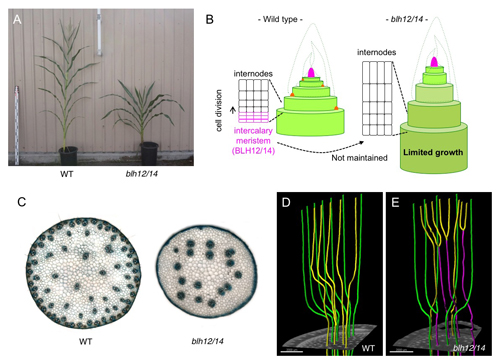
- (A) Wild-type (left) and blh12/14 (right) plants. blh12/14 double mutants show a dwarf phenotype.
- (B) Intercalary meristems are maintained by BLH12/14. In blh12/14, intercalary meristems were differentiated into internode and lost, leading to limited growth and dwarfness.
- (C) Transverse sections of wild-type (left) and blh12/14 double mutant (right) stems. blh12/14 had much fewer veins compared to wild type.
- (D) A vein network in the wild type stem extracted from micro CT data. Veins do not anastomose each other.
- (E) A vein network in the blh12/14 double mutant stem. Precociously anastomosed veins (magenta) were frequently observed.
Three new faculty have joined NIG as of April 1, 2017.
Three new faculty have joined NIG as of April 1, 2017.
professor
SAITO, Kuniaki : Invertebrate Genetics Laboratory
Associate Professor
KAWAMOTO, Shoko : Genetic Informatics Laboratory

- SAITO, Kuniaki
Professor

- KAWAMOTO, Shoko
Associate Professor
Assistant Professor












