Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Katsuhiko Minami in Genome Dynamics Laboratory won the “Student Presentation Award” and “wwPDB Student Award” at the 61st Annual Meeting of the Biophysical Society of Japan
Katsuhiko Minami, D5 student and JSPS Special Researcher in Genome Dynamics Laboratory, received the “Student Presentation Award” at the 61st Annual Meeting of the Biophysical Society of Japan held in Nagoya, Japan, on November 14th – 16th. In addition, He also received the “wwPBD Student Award”, which is given to the best presenters of research related to protein / nucleic acid structural studies.
- Awarded presentation title: Replication-dependent histone (Repli-Histo) labeling revealed that chromatin motion can determine DNA replication timing



Springer protocol “Zebrafish” (3rd edition) has been published.
Kawakami Group / Laboratory of Molecular and Developmental Biology
Zebrafish: Methods and Protocols (Third edition)
James F. Amatruda, Corinne Houart, Koichi Kawakami, Kenneth D. Poss
Methods in Molecular Biology (2023) 2707
The third edition of the zebrafish book is divided into four sections, reflecting the latest developments in zebrafish research. In Part 1, “Disease Models,” methods for modeling various human diseases using zebrafish are described. Part 2, “Neuroscience,” highlights the crucial role of fish models in the study of the vertebrate nervous system. Part 3, “Regeneration,” introduces tools and approaches using zebrafish to study stem cells and regenerative biology, which are increasingly important for human health. Part 4, “Genetics and Genomics,” details essential genetic and genomic methodologies for zebrafish research. From the Kawakami Laboratory, the following chapters were published:
Capter11: Pradeep Lal, Hideyuki Tanabe, and Koichi Kawakami. Genetic identification of neural circuits essential for active avoidance fear conditioning in adult zebrafish. Chapter 17: Kazuhide Asakawa, Hiroshi Handa, and Koichi Kawakami. In vivo optogenic phase transition of an intrinsically disordered protein. Chapter 20: Tomoya Shiraki and Koichi Kawakami. Generation of transgenic fish harboring CRISPR/Cas9-mediated somatic mutations via a tRNA-based multiplex sgRNA expression

“Metagenomic Thermometer” a novel approach to predict environmental temperatures based on metagenomic sequences.
Metagenomic Thermometer
Masaomi Kurokawa, Koichi Higashi, Keisuke Yoshida, Tomohiko Sato, Shigenori Maruyama, Hiroshi Mori, and Ken Kurokawa
DNA Research (2023) 30, dsad024 DOI:10.1093/dnares/dsad024
![]() Press release (In Japanese only)
Press release (In Japanese only)
Researchers at National Institute of Genetics have developed a groundbreaking tool, the “Metagenomic Thermometer,” which predicts environmental temperatures by analyzing the DNA of microorganisms in any given habitat. This innovative approach offers new insights into how environmental conditions influence life at a microbial level and could have wide-reaching implications in environmental science, biotechnology, and human health.
In a trailblazing study published in DNA Research, a team led by Professor Ken Kurokawa has unveiled the “Metagenomic Thermometer.” This tool marks a significant leap in environmental microbiology, enabling scientists to gauge the temperature of an environment by studying the genetic makeup of its microbial inhabitants.
The “Metagenomic Thermometer” operates on a groundbreaking principle: Professor Kurokawa’s team have extended the concept of estimating the optimal growth temperature (OGT) of a microorganism from the amino acid frequency of its genetic information, and developed a method of estimating the ambient temperature from the amino acid frequency in the collective genetic information of the microbial community present.
This revolutionary method was rigorously tested across diverse ecosystems, including hot springs, soil samples, and even the human gut. Remarkably, when applied to human gut metagenomic samples, the thermometer could accurately estimate human body temperature, underscoring the profound interplay between our internal environment and the microbial world.
Professor Kurokawa’s team believes that this tool doesn’t just measure temperature — it offers a new lens to understand how temperature drives the assembly of microbial communities. By focusing on amino acid composition rather than just microbial species, the Metagenomic Thermometer provides a more nuanced view of how life adapts to its environment.
The implications of this research are vast. From monitoring climate change impacts on microbial biodiversity to optimizing conditions for biotechnological applications, and even understanding human health in relation to our microbiome, the Metagenomic Thermometer stands as a testament to the power of innovative scientific inquiry.
“This research not only presents a novel tool but also opens up new avenues for understanding the intricate relationship between life and the environment,” said Professor Kurokawa. “We are excited about the potential applications of the Metagenomic Thermometer in various fields, including ecology, medicine, and biotechnology.”
- Variation in Gut Microbiome with Body Temperature:
The research proposes that body temperature significantly influences the gut microbiome composition. This revelation provides a deeper understanding of how bodily changes can impact gut health. - Predicting Deep Body Temperature:
Utilizing metagenomic analysis, the team has developed a ‘Metagenomic Thermometer’, which potentially allows for the estimation of human deep body temperature based on gut microbiota. This innovative approach offers a non-invasive method to gauge internal body temperature, a crucial health parameter. - Personalizing Medical Treatments:
The findings suggest exciting possibilities in tailoring treatments such as FMT and LBPs based on individual body temperatures. This could lead to more effective and personalized therapies for patients. - Probiotics Tailored to Body Temperature:
The research opens avenues for developing probiotics like yogurt and lactobacillus beverages specifically designed for individuals with varying body temperatures, enhancing their effectiveness. - Future Research Directions:
The team anticipates further exploration into how aging and diseases that lower body temperature affect gut microbiome composition. Additionally, the study’s methodologies could enhance predictions about bacterial community changes due to global warming, potentially influencing CO2 emission patterns.
For more information about this research, contact Office for Research Development.
– The study, “Metagenomic Thermometer,” was published in DNA Research.
– Interviews with the research team can be arranged upon request.
– High-resolution images and graphics explaining the Metagenomic Thermometer are available.
Contact:
Office for Research Development
National Institute of Genetics
National Institute of Genetics is a world-leading institution in genetics, genomics and bioinformatics. Our commitment to innovation and excellence places us at the forefront of global scientific endeavors.
Important notice regarding the entrance examination conducted in the winter of FY2023 (January 2024)
Taming the perils of photosynthesis – constraints on the evolution of the protective mechanisms
Miyagishima Group / Symbiosis and Cell Evolution Laboratory
Taming the perils of photosynthesis by eukaryotes: constraints on endosymbiotic evolution in aquatic ecosystems
Shin-ya Miyagishima
Communications Biology (2023) 6, 1150 DOI:10.1038/s42003-023-05544-0
An ancestral eukaryote acquired photosynthesis by genetically integrating a cyanobacterial endosymbiont as the chloroplast. The chloroplast was then further integrated into many other eukaryotic lineages through secondary endosymbiotic events of unicellular eukaryotic algae. While photosynthesis enables autotrophy, it also generates reactive oxygen species that can cause oxidative stress. To mitigate the stress, photosynthetic eukaryotes employ various mechanisms, including regulating chloroplast light absorption and repairing or removing damaged chloroplasts by sensing light and photosynthetic status. Recent studies have shown that, besides algae and plants with innate chloroplasts, several lineages of numerous unicellular eukaryotes engage in acquired phototrophy by hosting algal endosymbionts or by transiently utilizing chloroplasts sequestrated from algal prey in aquatic ecosystems. In addition, it has become evident that unicellular organisms engaged in acquired phototrophy, as well as those that feed on algae, have also developed mechanisms to cope with photosynthetic oxidative stress. These mechanisms are limited but similar to those employed by algae and plants. Thus, there appear to be constraints on the evolution of those mechanisms, which likely began by incorporating photosynthetic cells before the establishment of chloroplasts by extending preexisting mechanisms to cope with oxidative stress originating from mitochondrial respiration and acquiring new mechanisms.
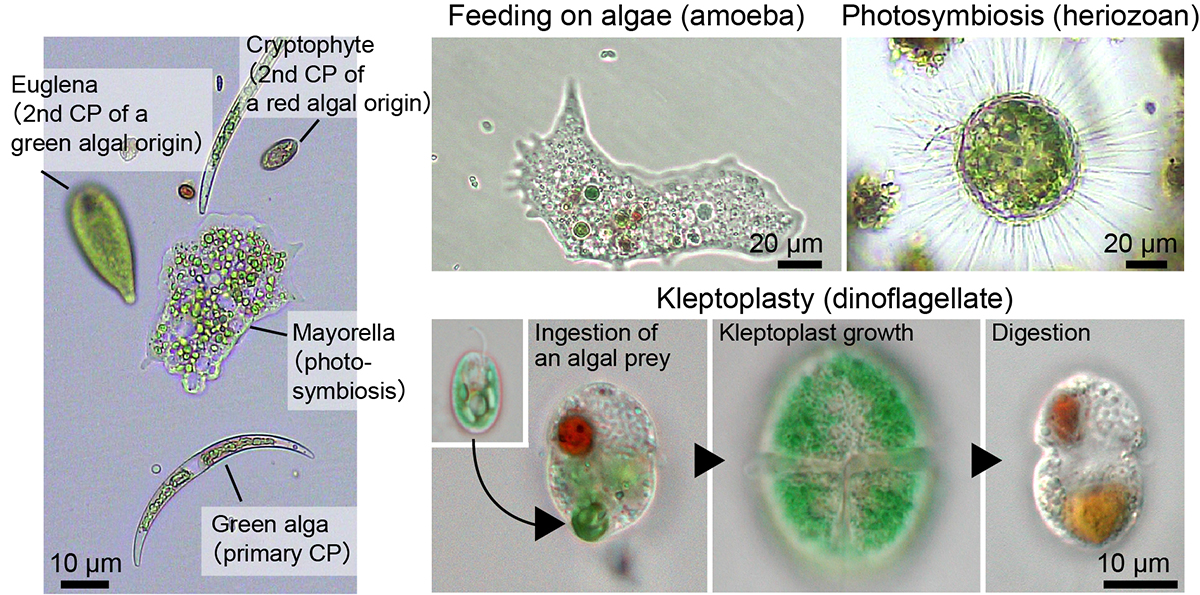
Figure: Examples of unicellular organisms with primary chloroplasts originating from cyanobacterial endosymbiosis, secondary chloroplasts from eukaryotic algal endosymbiosis, and organisms exhibiting phagotrophy, phototrophy, and kleptoplasty. Figure courtesy of Dr. Onuma (Kobe Univ.) and Mr. Okada (Nat. Inst. of Genet.).
A Practical Assembly Guideline for Genomes with Various Levels of Heterozygosity
Nakamura Group / Genome Informatics Laboratory
A Practical Assembly Guideline for Genomes with Various Levels of Heterozygosity
Takako Mochizuki, Mika Sakamoto, Yasuhiro Tanizawa, Takuro Nakayama, Goro Tanifuji, Ryoma Kamikawa, Yasukazu Nakamura*
*Corresponding Author
Briefings in Bioinformatics (2023) 24, bbad337 DOI:10.1093/bib/bbad337
The advancement of long-read sequencing technologies, exemplified by subreads of Pacific Biosciences, has significantly advanced our ability to reconstruct genome sequences. While these technologies can generate long reads, they are plagued by high sequence errors. To address these errors and strive to construct long, highly accurate contig sets, various de novo assemblers have been developed.
In de novo assembly of diploid genomes, the complexity increases with higher heterozygosity. Therefore, heterozygosity is a significant factor influencing the completeness of de novo assembly. However, systematic evaluations of de novo assemblers for diploid genomes with various heterozygosity levels have not been conducted.
In this study, using genomes with varying levels of heterozygosity, we conducted a series of processes, including estimation of genome characteristics such as genome size and heterozygosity, de novo assembly, polishing, and removing contigs including alleles. We have presented a guideline for constructing a representative haplotype set based on heterozygosity levels.
This work was supported by JSPS grant-in-aid for Scientific Research on Innovative Areas, Platform for Advanced Genome Science [16H06279], and KAKENHI [15H05606 and 19H03274, 20H03305, 17H03723].
Computations were partially performed on the National Institute of Genetics supercomputer.
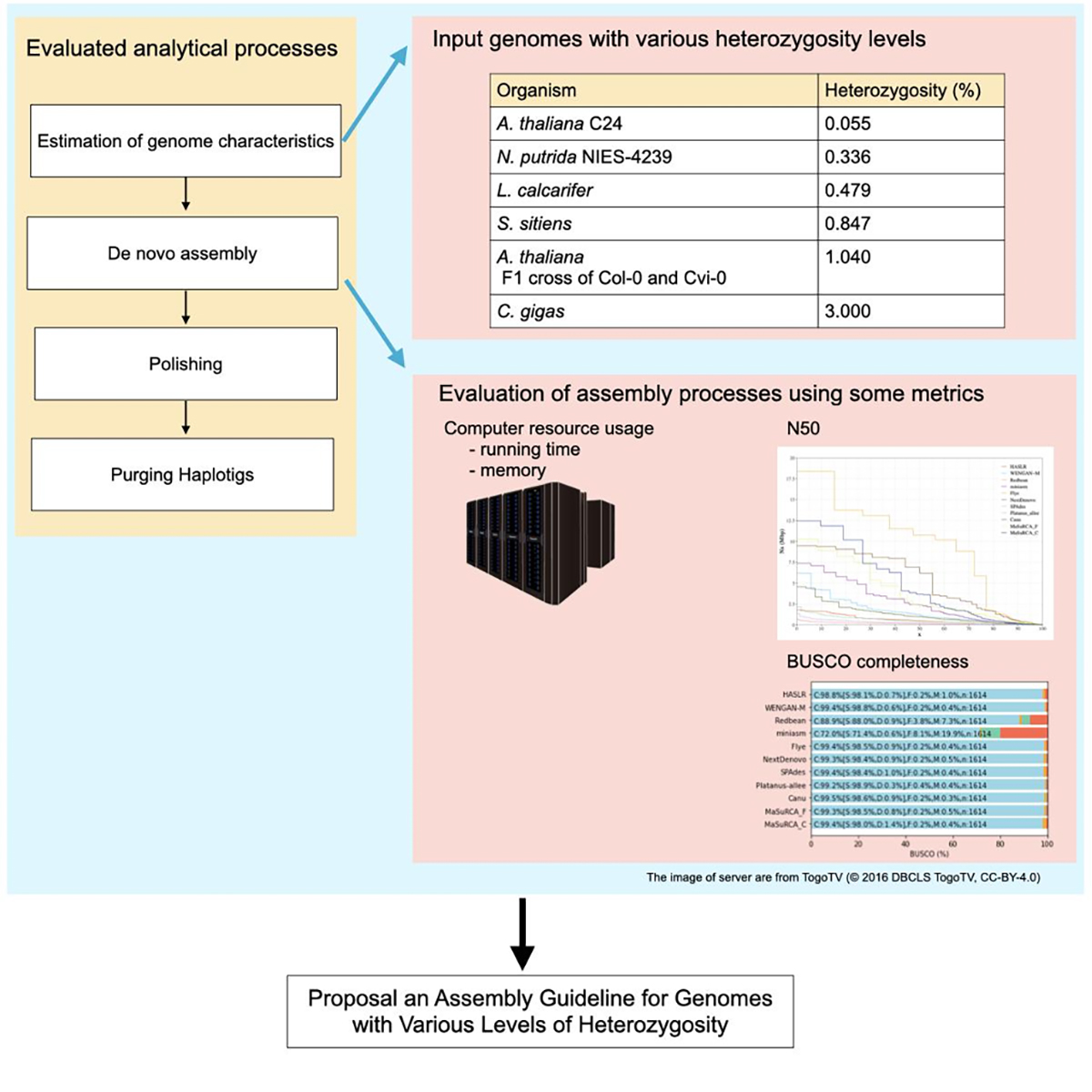
Figure: An evaluation process for genome assembly using genomes with various levels of heterozygosity
First Japanese record of freshwater prawn from Ryukyu Archipelago
Kitano Group / Ecological Genetics Laboratory
First record of Macrobrachium mammillodactylus (Thallwitz, 1891) (Crustacea, Decapoda, Palaemonidae) from Japan.
Yusuke Fuke, Tomoaki Maruyama.
Check List (2023) 19, 821-826 DOI:10.15560/19.6.821
The freshwater prawn genus Macrobrachium has more than 270 known species, with remarkably high species diversity in tropical freshwaters. Most of the widely distributed species in the Pacific have amphidromous migratory life history, which allows them to expand their range via ocean currents.
In this study, Yusuke Fuke, JSPS Postdoctoral Fellow at the National Institute of Genetics, and Tomoaki Maruyama of Trend Design Co., Ltd. identified the specimens collected from Miyako Island as Macrobrachium mammillodactylus (Thallwitz, 1891) based on genetic and morphological analysis. This marks the first record of this species in Japan and its northernmost distribution. This species has been previously reported from Australia to central Taiwan. The specimens collected in this study were adults, suggesting that they likely overwintered on the island, indicating an expansion of their range to the Ryukyu Archipelago.
Range expansion from Southeast Asia to the Ryukyu Archipelago and Honshu has been observed in other southern freshwater shrimp species. By accumulating fundamental range data, we may be able to identify and predict the factors contributing to the range expansion of amphidromous species.

Figure: Macrobrachium mammillodactylus collected from Miyako Island, Japan. Photo by T. Maruyama.
Summer Internship at NIG “NIGINTERN2024”
“SOKENDAI Award” awarded to Dr. Harsha Somashekar (Plant Cytogenetics Laboratory)

Dr. Harsha Somashekar of the Plant Cytogenetics Laboratory has received the 11th “SOKENDAI Award” of the Graduate University for Advanced Studies, SOKENDAI.
The SOKENDAI Award was established in (Academic Year) 2018 to recognize degree recipients who have conducted research worthy of commendation and reported their accomplishments in an outstanding doctoral thesis. Dr. Somashekar was presented with a certificate and a trophy by Dr. Nagata, President of SOKENDAI, on September 28, 2023 during the graduation ceremony. Dr. Somashekar has also been awarded the Genetics Program’s Morishima Award.
▶ thesis title : GLUCAN SYNTHASE-LIKE5 promotes anther callose deposition to maintain timely initiation and progression of meiosis in rice (Oryza sativa L.)
Comment from Dr. Harsha Somashekar
I embarked on a journey at NIG in 2018 with a dream – to become an independent researcher, contribute to the world of knowledge, and address real-world challenges. Today, five years later, that dream stands fulfilled. This achievement owes itself to the unwavering support and guidance I have received from NIG, my nurturing parent institute of SOKENDAI University.
NIG is an extraordinary institution. It not only imparts knowledge but also instills in its students a unique perspective for tackling real-world issues. The most valuable lesson I’ve learned here is the art of creating new knowledge and doing science in a right way. Thanks to the high-quality training programs and courses provided at NIG, I have gained invaluable knowledge and skills in conducting research.
I extend my heartfelt gratitude to NIG for affording me this remarkable opportunity. I also want to recognize the immensely talented faculty members who have been my mentors and guides on this doctoral journey. Their enduring support and mentorship have been instrumental in shaping me as a researcher.
Heat-shock inducible clonal analysis reveals the stepwise establishment of cell fates in the rice stem
Press release
Nonomura Group / Plant Cytogenetics Laboratory
Technical Section/ Cell Architecture Laboratory
Heat-shock inducible clonal analysis reveals the stepwise establishment of cell fates in the rice stem
Katsutoshi Tsuda, Akiteru Maeno, and Ken-Ichi Nonomura
Plant Cell 2023 Sep 27 DOI:10.1093/plcell/koad241
![]() Press release (In Japanese only)
Press release (In Japanese only)
The stem, composed of nodes and internodes, is the axis of shoots commonly seen among seed plants and is the most important target for regulating plant height in breeding practice. It is produced from the shoot apical meristem as a phytomer unit, along with the adjacent leaves and axillary buds. However, compared to other major organs such as leaves, roots, and flowers, our understanding of stem development has lagged far behind. Katsutoshi Tsuda, Akiteru Maeno, and Kenichi Nonomura at the National Institute of Genetics, Research Organization of Information and Systems, have defined the rice stem structure and described the developmental process using various approaches including micro CT. They also established a heat-shock inducible clonal analysis system and analyzed how cell fate establishment occurs for each organ during the development of the phytomer, which consists of the leaf, stem, and axillary bud. They found that cell fate establishment proceeds in a stepwise manner. First, cell fates for the phytomer are established prior to its initiation. Then, the emergence of cells that are destined for node splits of leaf and stem fates. Subsequently, the fate for the axillary bud fate is established. Finally, cell fate determination to the internodes occurs in a limited number of cells. This study clarified the key events and their chronological sequence in stem development, which had been unknown for a long time. It is expected to provide a basis for further research at the molecular level, which will lead to the improvement of stem traits for the design of ideal crops in the future.
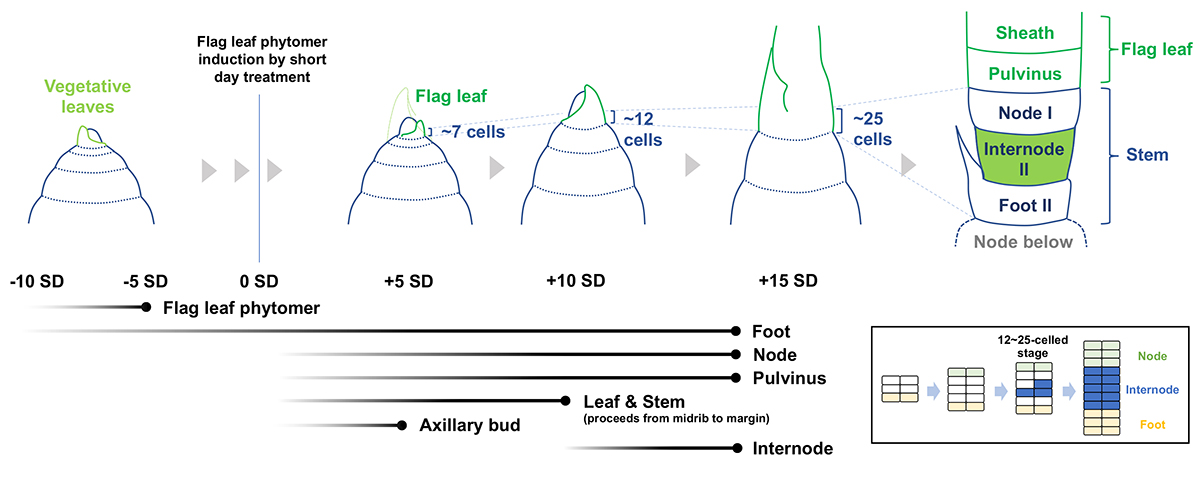
Figure: The temporal order of cell fate acquisition in the rice stem. The fate of the flag leaf phytomer is established 10 to 5 days prior to its initiation. Fates for non-elongating domains such as foot II, node I, and the pulvinus start being established earlier than elongating domains. Internode II is the last part of the stem whose cell fate determination starts. It is likely to originate from a very limited number of cells between node I and foot II (inset). Black gradient lines at the bottom left show the start (left) and end (right) of cell fate establishment. Stages at which the founder cells are fully present are indicated by dots at the end of black lines.
Genome and transcriptome analyses reveal genes involved in the formation of fine ridges on petal epidermal cells in Hibiscus trionum
Arita Group / Biological Networks Laboratory
Genome and transcriptome analyses reveal genes involved in the formation of fine ridges on petal epidermal cells in Hibiscus trionum
Shizuka Koshimizu*, Sachiko Masuda, Arisa Shibata, Takayoshi Ishii, Ken Shirasu, Atsushi Hoshino, Masanori Arita
* Corresponding author
DNA Research 2023 Sep 11 DOI:10.1093/dnares/dsad019
Hibiscus trionum, commonly known as the ‘Flower of an Hour’, is an easily cultivated plant in the Malvaceae family, widespread in tropical and temperate regions, including drylands. The purple base part of its petal exhibits structural color due to the fine ridges on the epidermal cell surface, and the molecular mechanism of ridge formation has been actively investigated. We performed genome sequencing of H. trionum using a long-read sequencing technology with transcriptome and pathway analyses to identify candidate genes for fine structure formation. The ortholog of AtSHINE1, which is involved in the biosynthesis of cuticular wax in Arabidopsis thaliana, was significantly overexpressed in the iridescent tissue. In addition, orthologs of AtCUS2and AtCYP77A, which contribute to cutin synthesis, were also overexpressed. Our results provide important insights into the formation of fine ridges on epidermal cells in plants using H. trionum as a model.
This study was conducted by a collaborative research group comprising the National Institute of Genetics (Shizuka Koshimizu and Masanori Arita), RIKEN (Sachiko Masuda, Arisa Shibata, and Ken Shirasu), Arid Land Research Center, Tottori Univ. (Takayoshi Ishii), and National Institute for Basic Biology (Atsushi Hoshino).
This work was supported by JSPS KAKENHI grants to KS (20H05909), JST ACT-X grant to SK (JPMJAX21B6), a grant from Sapporo Bioscience Foundation to SK, and NIBB Collaborative Research Program to SK (20-341, 21-223, 22NIBB308, and 23NIBB314).
Reference
Moyroud, E., Wenzel, T., Middleton, R., et al. 2017, Disorder in convergent floral nanostructures enhances signalling to bees. Nature (2017) 550, 469–474.
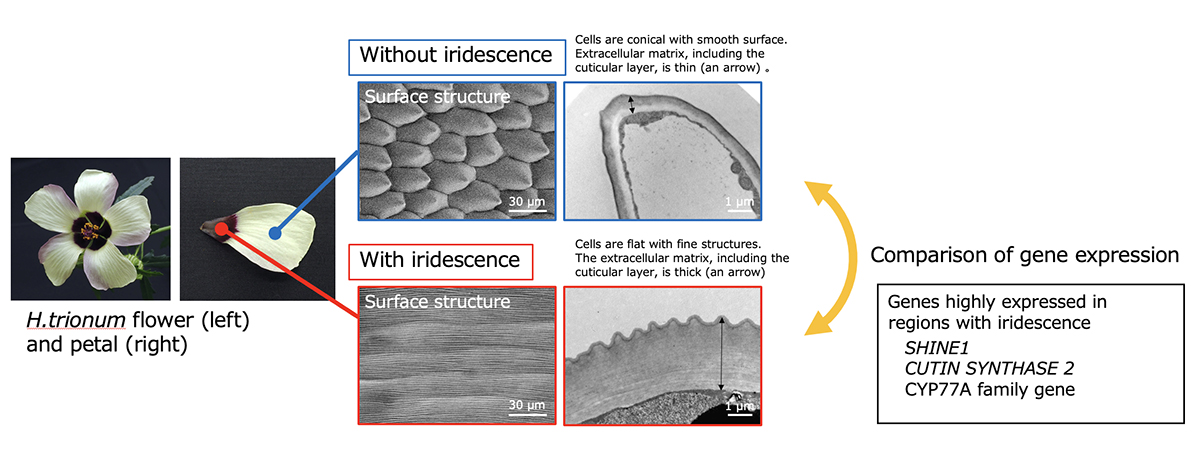
Figure: H. trionum flower and comparison of the surface structures
Mr. Harsha Somashekar (Plant Cytogenetics Laboratory) received the Morishima Award

On September 12, 2023, Mr. Harsha Somashekar (Plant Cytogenetics Laboratory, Nonomura Laboratory) received the Morishima Award from the Genetics Program, SOKENDAI. The Morishima Award is given to students in the Genetics Program to honor their outstanding performances during PhD studies and to encourage further achievements.
・Harsha Somashekar (Plant Cytogenetics Laboratory)
thesis title : GLUCAN SYNTHASE-LIKE5 promotes anther callose deposition to maintain timely initiation and progression of meiosis in rice (Oryza sativa L.)
Identifying a protein essential for replication initiation
Kanemaki Group / Molecular Cell Engineering Laboratory
In silico protein interaction screening uncovers DONSON’s role in replication initiation
Yang Lim, Lukas Tamayo-Orrego, Ernst Schmid, Zygimante Tarnauskaite, Olga V. Kochenova, Rhian Gruar, Sachiko Muramatsu, Luke Lynch, Aitana Verdu Schlie, Paula L. Carroll, Gheorghe Chistol, Martin A. M. Reijns, Masato T. Kanemaki, Andrew P. Jackson, and Johannes C. Walter
Science 2023 Aug 17 DOI:10.1126/science.adi3448
For cellular proliferation, it is essential to replicate chromosomal DNA that encodes genetic information. Until now, research on eukaryotic DNA replication has advanced using budding yeast as a model. In budding yeast, all the proteins required for replication initiation have been identified, and reconstitution experiments using these proteins have been conducted. In this reaction, assembling the CMG replicative helicase, composed of CDC45, MCM2–7, and GINS, is a crucial regulatory point. The Sld2 protein, discovered by Professor Emeritus Hiroyuki Araki at National Institute of Genetics, forms a pre-loading complex (pre-LC) with GINS, Polε, and Dpb11 and brings GINS to MCM2–7.
The formation of the CMG helicase is crucial in vertebrate cells as well. However, the proteins involved in CMG formation have not been fully identified. In this paper, we reported that DONSON, encoded by a causative gene for microcephaly, plays an essential role in the assembly of the CMG helicase by forming a pre-LC with GINS, Polε, and TOPBP1 (the Dpb11 homolog). In this study, we employed AI utilizing Alfafold2 multimer to predict protein-protein interactions and analyzed the function of DONSON in frog egg extracts, human cultured cells, and mice.
Scientific Officer Sachiko Sakamoto (Muramatsu) and Professor Masato Kanemaki generated human cells in which DONSON can be rapidly degraded with AID2. This research was carried out as an international collaboration with Professor Andrew Jackson at University of Edinburgh and Professor Johannes Walter at Harvard University.
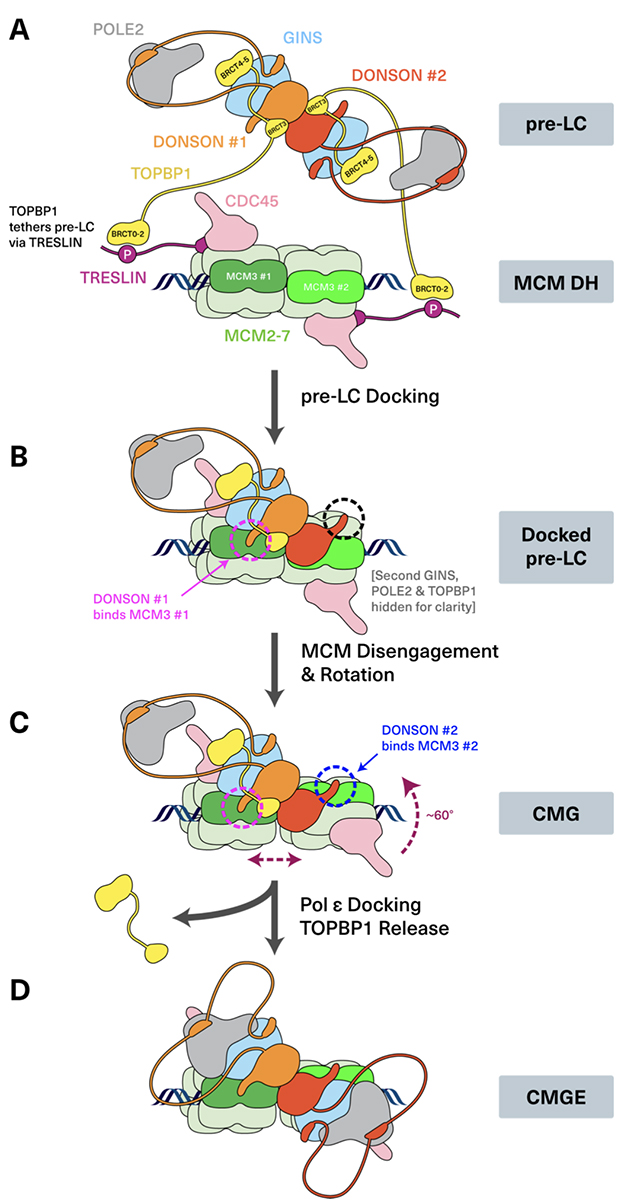
Figure: A model of the CMG helicase assembly in vertebrate cells.
Uncovering how the Golgi apparatus impacts early postnatal neuron development
Press release
Golgi polarity shift instructs dendritic refinement in the neonatal cortex by mediating NMDA receptor signaling
Naoki Nakagawa, Takuji Iwasato
Cell Reports 2023 Jul 28 DOI:10.1016/j.celrep.2023.112843
![]() Press release (In Japanese only)
Press release (In Japanese only)
EurekAlert! link about this artcle
Neurons are the cells that constitute neural circuits and use chemicals and electricity to receive and send messages that allow the body to do everything, including thinking, sensing, moving, and more. Neurons have a long fiber called an axon that sends information to the subsequent neurons. Information from axons is received by branch-like structures that fan out from the cell body, called dendrites.
Dendritic refinement is an important part of early postnatal brain development during which dendrites are tailored to make specific connections with appropriate axons. In a recently published paper, researchers present evidence showing how a mechanism within the neurons of a rodent involving the Golgi apparatus initiates dendritic refinement with the help of the neuronal activity received by a receptor of a neurotransmitter called N-methyl-D-aspartate-type glutamate receptor (NMDAR).
The paper was published in Cell Reports on July 28. Read more>
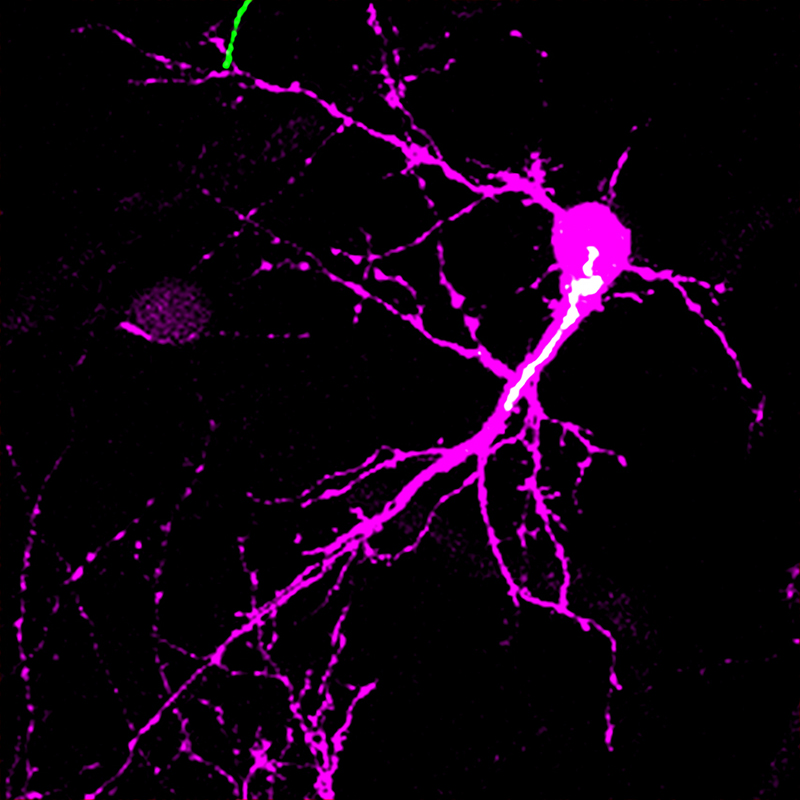
Figure: The image shows a spiny stellate neuron (magenta) and its Golgi apparatus (white) in the mouse barrel cortex at postnatal day 5. The study found that the Golgi apparatus shows a biased distribution in one side of the neuron only during the postnatal critical period of circuit reorganization. This lateral Golgi polarity instructs the direction in which the neuron expands its dendrites for establishing precise neuronal circuit.
A potential key mechanism underlying ALS: Excessive TDP-43 protein decreases motor neuron excitability.
Dysregulated TDP-43 proteostasis perturbs excitability of spinal motor neurons during brainstem-mediated fictive locomotion in zebrafish
Kazuhide Asakawa, Hiroshi Handa, Koichi Kawakami
evelopment, Growth & Differentiation 2023 July 15 DOI:10.1111/dgd.12879
Amyotrophic lateral sclerosis (ALS) is a debilitating disease that causes muscle weakness and eventually fatal paralysis. A major hallmark of ALS is the deposition of the aggregate form of TDP-43 protein in motor neurons, a type of nerve cell that directs muscle contractions. This motor neuron pathology can occur without genetic mutations in the coding sequence of the TDP-43-encoding gene TARDBP. Whether and how wild-type TDP-43 drives pathological changes in motor neurons remain largely unexplored.
In the present study, Asakawa and colleagues used calcium imaging to directly compare the neural activity of motor neurons with and without excessive TDP-43 protein during fictive locomotion. They found that excessive amounts of TDP-43 protein reduced the excitability of motor neurons (Figure). This finding suggests that the reduced excitability caused by excessive TDP-43 potentially contributes to asymptomatic pathological lesions of motor neurons and movement disorders in patients with ALS.
This work was published in the journal Development Growth & Differentiation on the 15th of July, 2023. The study was conducted by a collaborative research group comprising the National Institute of Genetics (Kazuhide Asakawa and Koichi Kawakami) and Tokyo Medical University (Hiroshi Handa).
This work was supported by the Nakabayashi Trust For ALS Research (K.A.), The Kato Memorial Trust For Nambyo Research (K.A.), Daiichi-Sankyo Foundation of Life Science (K.A.), Takeda Science Foundation (K.A.), KAKENHI grant numbers JP16K07045 (K.A.), JP19K06933 (K.A.), JP22H02958 (K.A.), JP23H04266 (K.A.), JP21H02463 (K.K.) and AMED-PRIME grant number JP23gm6410011h0003 (K.A.), and the National BioResource Project (NBRP) (K.K.).
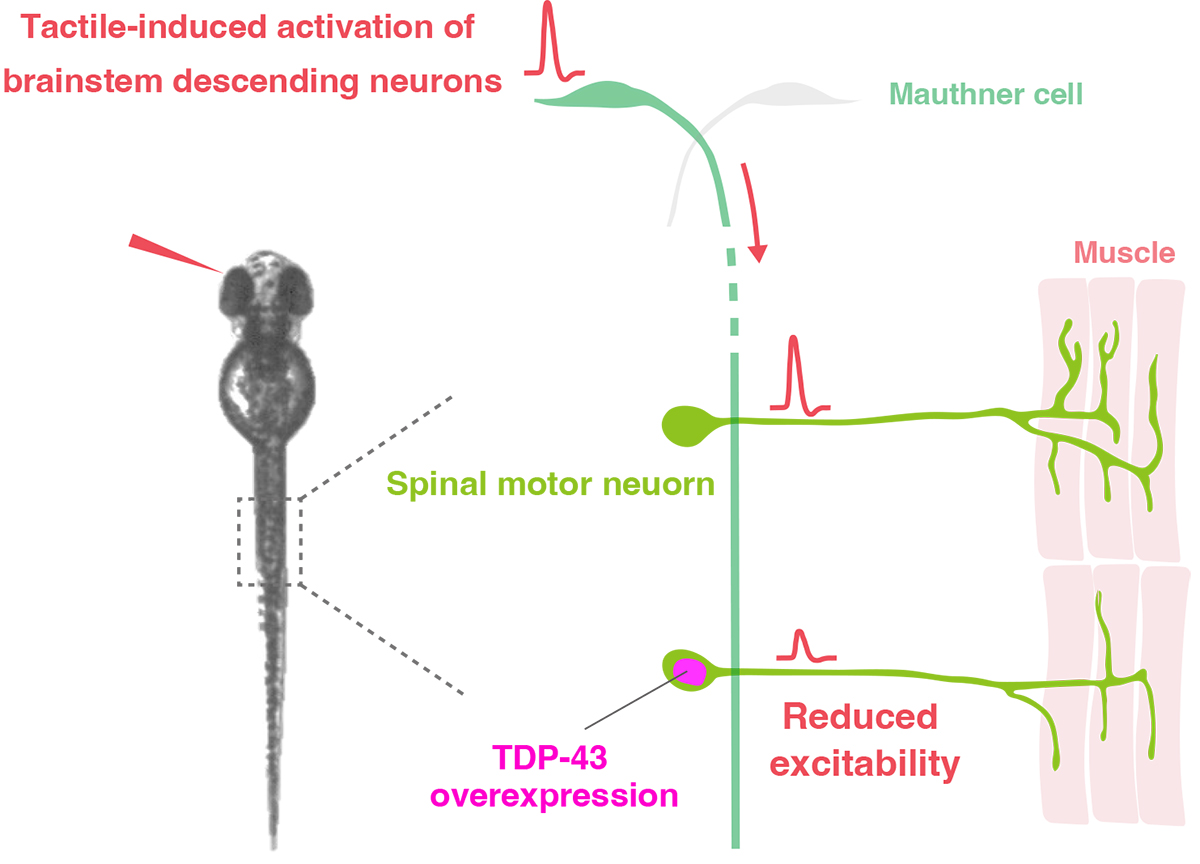
Figure: Excessive amounts of TDP-43 protein reduce motor neuron excitability
Summer Holiday (Aug. 15-16)
NIG will be closed from August 15 to August 16, 2023 for summer holiday.
Thank you for your understanding and cooperation.















