Development of fast and accurate taxonomic assignment tool VITCOMIC2
VITCOMIC2: visualization tool for the phylogenetic composition of microbial communities based on 16S rRNA gene amplicons and metagenomic shotgun sequencing.
Mori H, Maruyama T, Yano M, Yamada T, Kurokawa K. BMC Syst Biol
BMC Systems Biology 12 (Suppl 2), 30, 2018. DOI:10.1186/s12918-018-0545-2
Dr. Hiroshi Mori and Dr. Ken Kurokawa in the Genome Evolution laboratory from the Center for Information Biology in National Institute of Genetics, and colleagues developed a web application “VITCOMIC2” to infer taxonomic composition of a microbial community from the metagenomic sequencing data and the 16S rRNA gene amplicon sequencing data. VITCOMIC2 conducts fast and accurate taxonomic assignment of metagenomic data by using a GPU-based DNA sequence similarity search tool CLAST. Using VITCOMIC2, users can avoid the taxonomic assignment ambiguities which are derived from sequence clustering before conducting taxonomic assignment. VITCOMIC2 can be used in http://vitcomic.org .
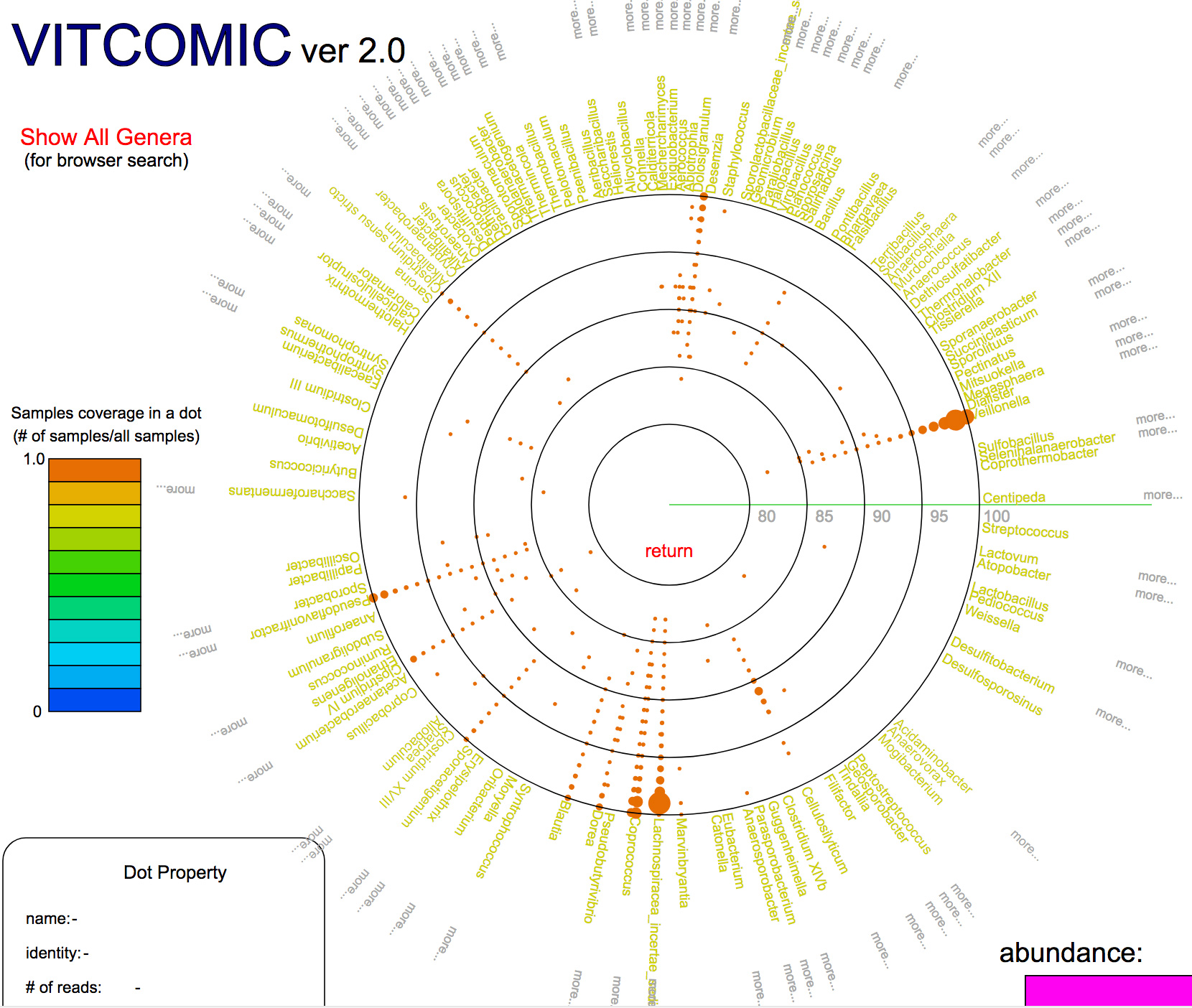
図:Example of a VITCOMIC2 result















