Mori Group • Genome Diversity Laboratory
Genome biology to understand the organismal diversity in environments
Faculty
Research Summary
Organisms inhabit various environments and exhibit remarkable genomic diversity. Our main research goal is to understand the relationships between genome diversity and habitat diversity. We use various bioinformatics and statistical methodologies related to comparative genomics and metagenomics in our research. Our research is related to ancient DNA analysis of extinct animals, and genome reconstruction of phylogenetically novel bacteria from metagenomic sequencing data. We are also developing some bioinformatics methods and tools for genomics and metagenomics, and applying these methods to various collaborative research in the Advanced Genomics Center.
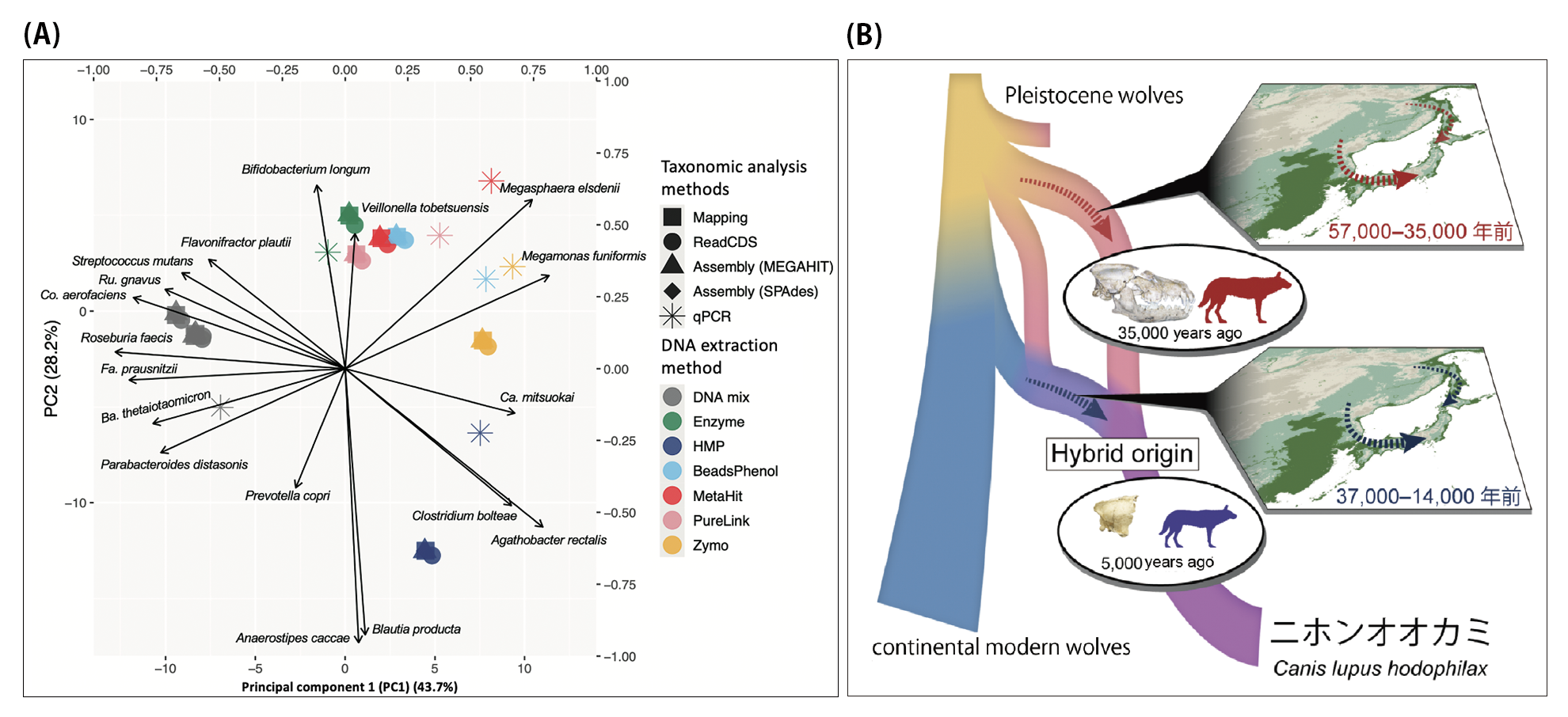
(B) The inferred history of the origin of the Japanese wolf. Our ancient DNA study of Pleistocene wolf remains recovered in Japan reveals that the origin of Japanese wolf is a hybrid of two morphologically distinct wolf lineages.
Selected Publications
Mori H, Ishikawa H, Higashi K, Kato Y, Ebisuzaki T, Kurokawa K. PZLAST: an ultrafast amino acid sequence similarity search server against public metagenomes. Bioinformatics. 2021 Jul 7;37(21):3944–6.
Segawa T, Yonezawa T, Mori H, Akiyoshi A, Allentoft ME, Kohno A, Tokanai F, Willerslev E, Kohno N, Nishihara H. Ancient DNA reveals multiple origins and migration waves of extinct Japanese brown bear lineages. R Soc Open Sci. 2021 Aug 4;8(8):210518.
Segawa T, Yonezawa T, Mori H, Kohno A, Kudo Y, Akiyoshi A, Wu J, Tokanai F, Sakamoto M, Kohno N, Nishihara H. Paleogenomics reveals independent and hybrid origins of two morphologically distinct wolf lineages endemic to Japan. Curr Biol. 2022 Jun 6;32(11):2494-2504.e5.
Mori H, Kato T, Ozawa H, Sakamoto M, Murakami T, Taylor TD, Toyoda A, Ohkuma M, Kurokawa K, Ohno H. Assessment of metagenomic workflows using a newly constructed human gut microbiome mock community. DNA Res. 2023 Jun 1;30(3):dsad010.
















