Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Efficient production of recombinant proteins using the Tol2 transposon
Efficient production of recombinant proteins in suspension CHO cells culture using the Tol2 transposon system coupled with cycloheximide resistance selection.
Keina Yamaguchi, Risa Ogawa, Masayoshi Tsukahara and Koichi Kawakami Scientific reports (2023) 13, 7628 DOI:10.1038/s41598-023-34636-4DNA recombination techniques in mammalian cells has been applied to the production of therapeutic proteins for several decades. To be used for commercial production, established cell lines should stably express target proteins with high productivity and acceptable quality for human use. In the conventional transfection method, the screening process is laborious and time-consuming since superior cell lines had to be selected from an enormous number of transfected cell pools and clonal cell lines with a wide variety of transgene insertion locations. In this study, we demonstrated that the combination of a Tol2 transposon system and cell selection by cycloheximide resistance is an efficient method to express therapeutic proteins, such as human antibody in suspension culture of Chinese hamster ovary cells. The resulting stable cell lines showed constant productivity and cell growth over long enough cultivation periods for recombinant protein production. We anticipate that this approach will prove widely applicable to protein production in research and development of pharmaceutical products.
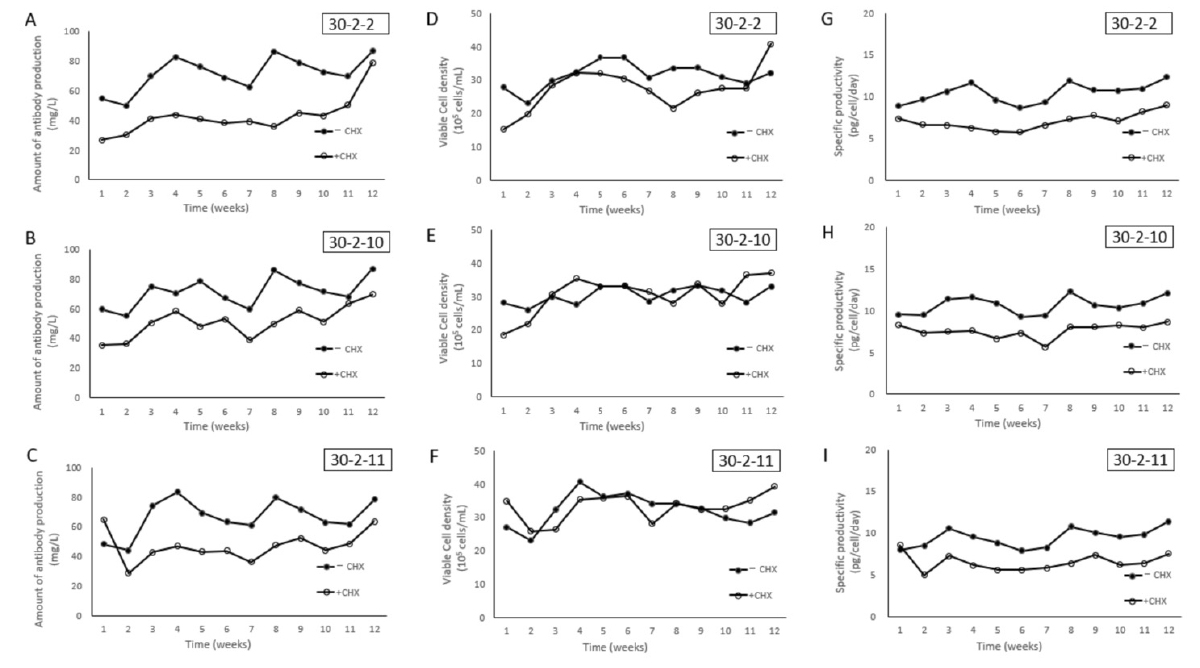
Figure: Antibody productivity and cell growth of clonal cell lines 30-2-2, 30-2-10, 30-2-11 in the presence or absence of CHX selection. Antibody productivity (A, B and C), cell growth (D, E and F) and specific antibody productivity (pg/cell/day) (G, H and I) in 30-2-2, 30-2-10, 30-2-11 were measured in the presence (open circles) or absence (black circles) of CHX selection pressure.
Guideline for Additional Application for 2023 NIG-JOINT (Joint Research-(A)) (Application was closed)
Is euchromatin really open in the cell?
Is euchromatin really open in the cell?
Kazuhiro Maeshima*#, Shiori Iida*, Masa A. Shimazoe, Sachiko Tamura, Satoru Ide
*cofirst authors; #corresponding author
Trends in Cell Biology 2023 June 27 DOI:10.1016/j.tcb.2023.05.007
The human genome chromatin can be classified into euchromatin and heterochromatin, which have high and low transcription activities, respectively. In the classical view, it was believed that euchromatin has an open and decondensed structure, while heterochromatin is highly condensed.
A research team led by Professor Kazuhiro Maeshima of Genome Dynamics Laboratory (NIG), including a graduate student (JSPS Research Fellow DC2) Shiori Iida, SOKENDAI graduate student Masa A. Shimazoe, Technical Stuff Sachiko Tamura, and Assistant Professor Satoru Ide have put forward the model that euchromatin essentially forms condensed domains with sizes ranging from 100-300 nm in diameter based on new evidence from genomics and advanced imaging studies. They discuss this novel view of euchromatin in the cell and how the revealed organization is relevant to genome functions.
Physically condensed domains provide a higher-order regulation of transcription, while the extended fiber loops do not. Condensed domains likely hinder the accessibility of large transcription complexes to their target sequence located in the inner core of chromatin domains.
This exclusion effect can repress unintended gene expression. Although such a core of condensed domains seems to be inaccessible, condensed domains have the liquid-like property, allowing small transcription factors to penetrate the core of domains. Such small transcription factors may help a target sequence to float up to the domain surface to be transcribed. The authors suggest that condensed chromatin domains can work as Lego blocks of mitotic chromosomes during cell division, which create a smoother process for chromosome assembly and disassembly.
This work was supported by JSPS Fellowship, JSPD grants (JP21H02453, JP22H05606, JP21H02535, JP20H05936, JP16H06279(PAGS)), JSPS Research Fellow (JP23KJ0996(DC2)), JST SPRING (JPMJSP2104).
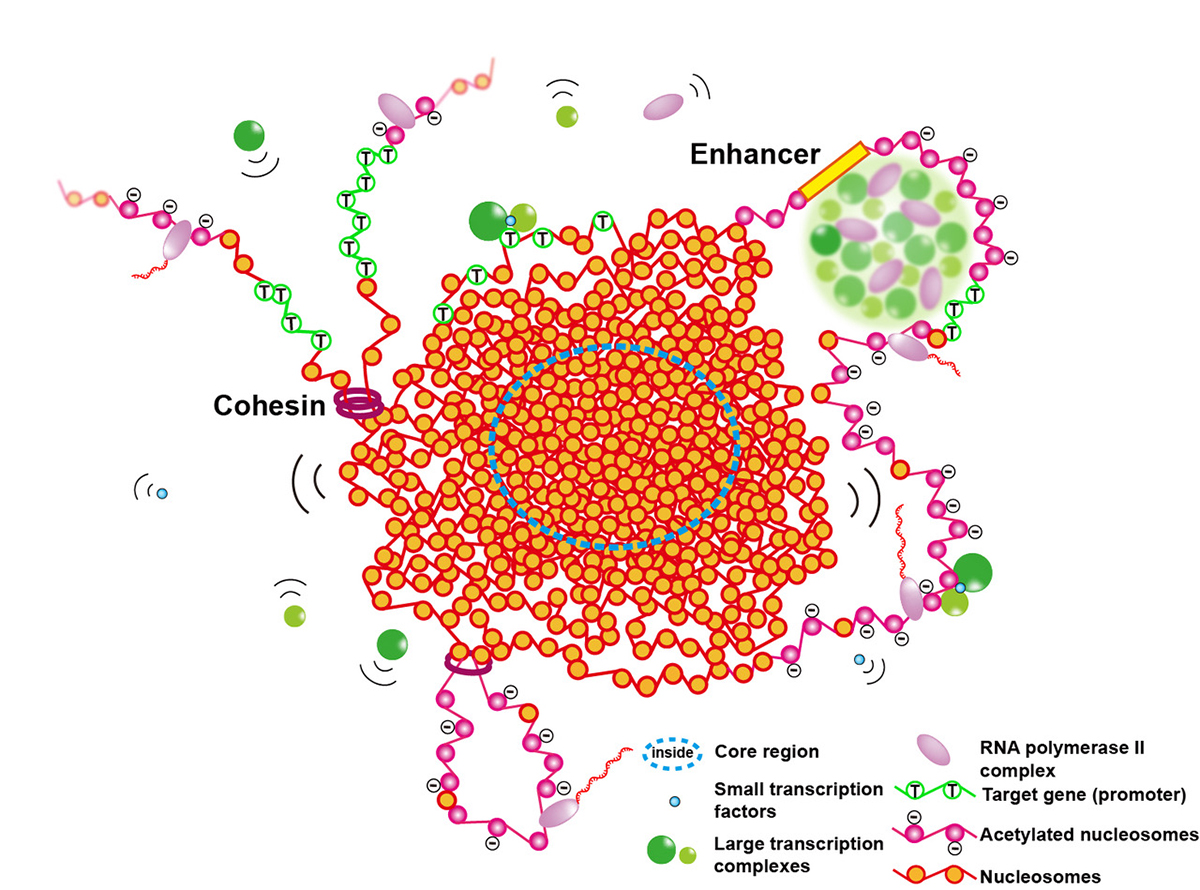
Figure: Euchromatic condensed domains provide higher-order regulation of transcription by physically excluding large transcription complexes (green spheres) from the inner core of the domains. Liquid-like property inside a condensed domain gives a certain degree of accessibility for gene expression. Small transcription factors interact with the target gene (open green circle nucleosomes) inside the inner core and cause it to float up to the domain surface to be transcribed. The condensed chromatin domains can work as Lego blocks of mitotic chromosomes during cell division.
NIG-GS (NIG Global Scholar) selection
事務職員1名及び施設系職員(電気又は機械)1名を募集します。
第一次試験「東海・北陸地区国立大学法人等職員採用試験」
第一次試験 7月 2日(日)
第一次試験合格者発表 7月20日(木)9時30分
■機関説明会■
第一次試験に合格された方について、以下のとおりオンラインにて機関説明会を実施します。
(実 施 日) 7月27日(木)13:30~(30分程度)
(内 容) オンラインにて機関の紹介、質疑応答を行います。
(参加方法) ご希望者は、7月26日(水)正午までに、以下URLよりお申込みください。
https://forms.gle/yEDBvqtVPdCDekQe6
*返信にて、Zoom情報をお知らせいたしますが、届かない場合には、以下担当までご連絡願います。
第二次試験(1次選考・最終選考) ※ 新型コロナウイルス感染症の状況によっては、実施方法を
オンライン面接などに変更する場合があります。
■1次選考■
(対象者) 令和5年度東海・北陸地区国立大学法人等職員採用試験(事務、電気及び機械)の第一次試験合格者
(日 時) 令和5年8月7日(月) 予備日:9日(水)
(会 場) 情報・システム研究機構国立遺伝学研究所 本館2階応接室
(https://www.nig.ac.jp/nig/ja/about-nig/access_ja)
(実施方法) 個別面接
(事前に送付する物) ・訪問カード(履歴、志望動機等)
※所定の様式をダウンロードして記入の上、8月4日(金)10時までに、下記申込み先まで送付願います。
(持参する物) ・第一次試験合格通知のメール(写)
■最終選考■
(対象者) 1次選考の合格者
(日 時) 令和5年8月31日(木)から9月1日(金)
(会 場) 情報・システム研究機構本部事務局
東京都港区虎ノ門4-3-13ヒューリック神谷町ビル2階
(https://www.rois.ac.jp/about/access.html)
※最終選考は、情報・システム研究機構本部で実施します。
【 申 込 方 法 】
令和5年8月2日(水)15:00までに
「受験番号、氏名、メールアドレス、電話番号」を人事・労務係(
*面接にお越しいただく交通費は、自己負担となりますのでご了承願います。
問い合せ先:管理部総務企画課人事・労務係(担当:鈴木)
TEL: 055-981-6716 E-mail:
POLR1A variant- and tissue-specific effects underlie phenotypic heterogeneity in craniofacial, neural and cardiac anomalies
POLR1A variant- and tissue-specific effects underlie phenotypic heterogeneity in craniofacial, neural and cardiac anomalies
Kelly Smallwood, Kristin E.N. Watt, Satoru Ide, Kristina Baltrunaite, Chad Brunswick, Katherine Inskeep, Corrine Capannari, Margaret P. Ada, Amber Begtrup, Debora R. Bertola, Laurie Demmer, Erin Demo, Orrin Devinsky, Emily R. Gallagher, Maria J. Guillen Sacoto, Robert Jech, Boris Keren, Jennifer Kussmann, Roger Ladda, Lisa A. Lansdon, Sebastian Lunke, Anne Mardy, Kirsty McWalters, Richard Person, Laura Raiti, Noriko Saitoh, Carol J. Saunders, Rhonda Schnur, Matej Skorvanek, Susan L. Sell, Anne Slavotinek, Bonnie R. Sullivan, Zornitza Stark, Joseph D. Symonds, Tara Wenger, Sacha Weber, Sandra Whalen, Susan M. White, Juliane Winkelmann, Michael Zech, Shimriet Zeidler, Kazuhiro Maeshima, Rolf W. Stottmann, Paul A Trainor, K. Nicole Weaver
The American Journal of Human Genetics (2023) 110, 809-825 DOI:10.1016/j.ajhg.2023.03.014
Heterozygous pathogenic variants in POLR1A, which encodes the largest subunit of RNA Polymerase I, were previously identified as the cause of Acrofacial Dysostosis, Cincinnati-type. The predominant phenotypes observed in the initial cohort of 3 individuals were craniofacial anomalies reminiscent of Treacher Collins syndrome. We subsequently identified 17 additional individuals with 12 unique (11 novel) heterozygous variants in POLR1A and observed numerous additional phenotypes including neurodevelopmental abnormalities and structural cardiac defects, in combination with highly prevalent craniofacial anomalies and variable limb defects. To understand the pathogenesis of this pleiotropy, we modeled an allelic series of POLR1A variants in vitro and in vivo. In vitro assessments demonstrate variable effects of individual pathogenic variants on ribosomal RNA synthesis and nucleolar morphology which supports the possibility of variant-specific phenotypic effects in patients. To further explore variant-specific effects in vivo, we used CRISPR/Cas9 gene editing to recapitulate two human variants in mice. Additionally, spatiotemporal requirements for Polr1a in developmental lineages contributing to congenital anomalies in patients were examined via conditional mutagenesis in neural crest cells (face and heart), the second heart field (cardiac outflow tract and right ventricle), and forebrain precursors in mice (Figure A). Consistent with its ubiquitous role in the essential function of ribosome biogenesis, we observed that loss of Polr1a in any of these lineages causes death of the affected cells and resulting embryonic malformations. Altogether, our work greatly expands the phenotype of human POLR1A-related disorders and demonstrates variant-specific effects and differential tissue-specific requirements that provide novel insights into the underlying pathogenesis of ribosomopathies.
Assistant Professor Satoru Ide and Professor Kazuhiro Maeshima of Genome Dynamics Laboratory performed in vitro assessment of POLR1A variants, which impact rRNA transcription and the nucleolar morphology in human culture cells expressing wild-type or mutant POLR1A. Using a machine learning algorithm called “wndchrm”, Ide and Maeshima found that nucleolar morphologies of the mutants (based on POLR1A foci) are somehow corelated with their rRNA transcription capacities or status (Figure B).
The part to which NIG contributed was supported by the Japan Society for the Promotion of Science (JSPS) grants (22H05606, 21H02535 to SI; 19H05273 ,20H05936 to KM) etc. The image analysis by the machine learning algorithm “wndchrm” was conducted in collaboration with Dr. Noriko Saito in The Cancer Institute of JFCR.
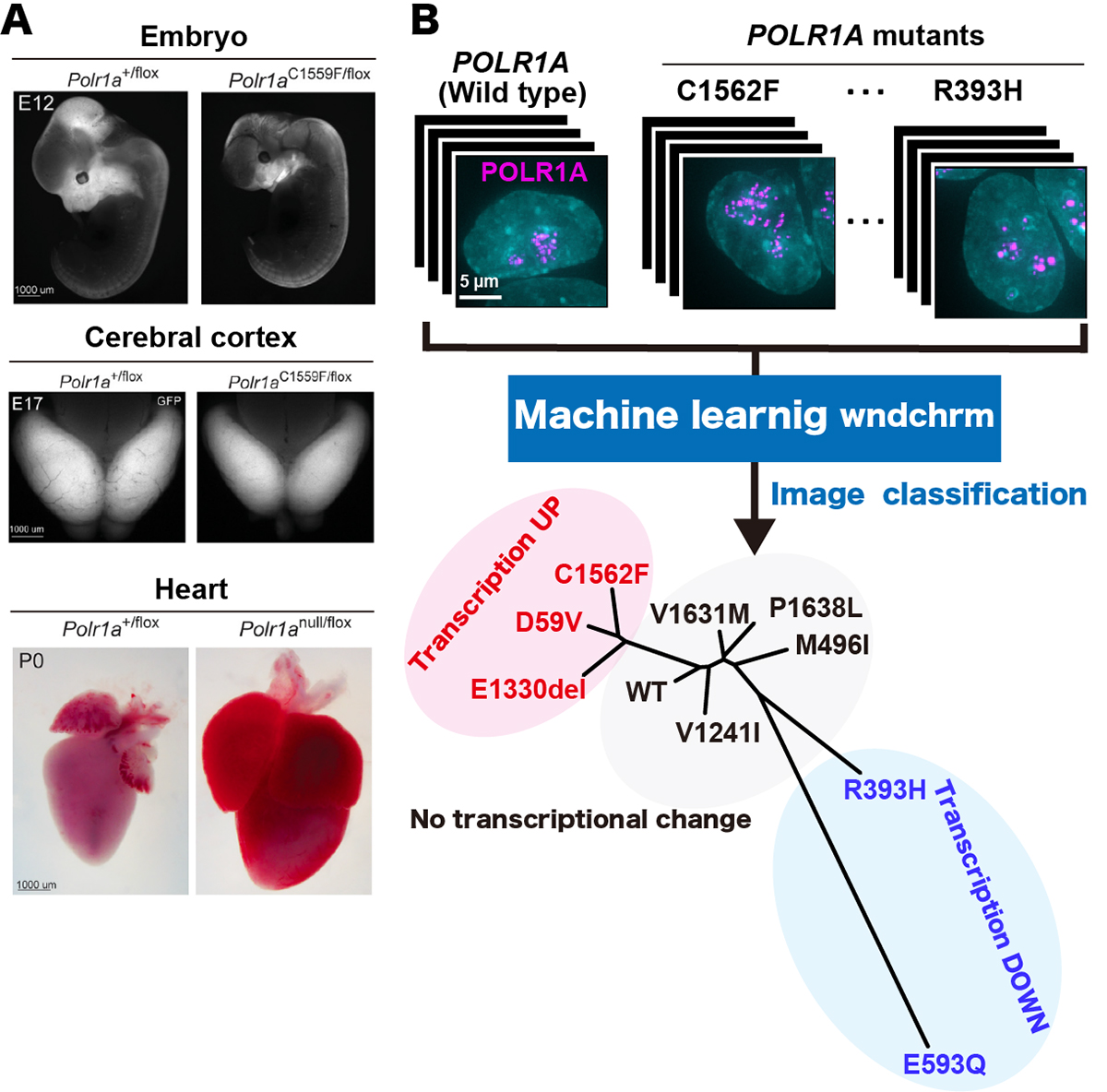
Figure: (A)Polr1a variant murine embryos demonstrate more hypoplastic craniofacial primordia at E12 (upper panel, right), a reduced cerebral cortex area at E12 (middle panel, right) and gross enlargement of heart at P0 (lower panel, right), compared to wild type (all the panels, left), respectively. (B) (upper panel) Localization of fluorescently labeled POLR1A (cellular nucleus, cyan and human POLR1A variants, magenta). (lower panel) Phylogeny shows the similarity of the nucleolar morphologies of wild type and nine POLR1A variants. POLR1A variants showing transcription upregulation, transcription downregulation and no transcriptional change are indicated in red, blue, and black, respectively.
Assessment of metagenomic workflows using a newly constructed human gut microbiome mock community
![]()
Assessment of metagenomic workflows using a newly constructed human gut microbiome mock community
Hiroshi Mori*, Tamotsu Kato*, Hiroaki Ozawa, Mitsuo Sakamoto, Takumi Murakami, Todd D. Taylor, Atsushi Toyoda, Moriya Ohkuma, Ken Kurokawa, Hiroshi Ohno
*equal contribution
DNA Research 2023 May 31 DOI:10.1093/dnares/dsad010
Press release (In Japanese only)
To quantify the biases introduced during human gut microbiome studies, analyzing an artificial mock community as the reference microbiome is indispensable. However, there are still limited resources for a mock community which well represents the human gut microbiome. Here, we constructed a novel mock community comprising the type strains of 18 major bacterial species in the human gut and assessed the influence of experimental and bioinformatics procedures on the 16S rRNA gene and shotgun metagenomic sequencing. We found that DNA extraction methods greatly affected the DNA yields and taxonomic composition of sequenced reads, and that some of the commonly used primers for 16S rRNA genes were prone to underestimate the abundance of some gut commensal taxa such as Erysipelotrichia, Verrucomicrobiota and Methanobacteriota. Binning of the assembled contigs of shotgun metagenomic sequences by MetaBAT2 produced phylogenetically consistent, less-contaminated bins with varied completeness. The ensemble approach of multiple binning tools by MetaWRAP can improve completeness but sometimes increases the contamination rate. Our benchmark study provides an important foundation for the interpretation of human gut microbiome data by providing means for standardization among gut microbiome data obtained with different methodologies and will facilitate further development of analytical methods.
Source: Hiroshi Mori et al., DNA Research doi: 10.1093/dnares/dsad010
Shiori Iida in Genome Dynamics Laboratory has completed Analytical and Quantitative Light Microscopy course at Marine Biological Laboratory

She is wearing a T-shirt of Analytical and
Quantitative Light Microscopy 2023 with
an illustration of a lobster and a microscope.
Shiori Iida, a D4 student and JSPS research fellow DC2 at Genome Dynamics Laboratory, has successfully completed the Analytical and Quantitative Light Microscopy held at Marine Biological Laboratory in Woods Hole, United States, from April 26th to May 5th, 2023. This course is a historic and educational program focused on light microscopy, held annually at the Marine Biological Laboratory.
The course fee and travel expenses were supported by Morishima Fund of the Genetics course at SOKENDAI.
The review: Genetic Regulation of Mitosis–Meiosis Fate Decision in Plants
Genetic Regulation of Mitosis–Meiosis Fate Decision in Plants: Is Callose an Oversighted Polysaccharide in These Processes?
Harsha Somashekar and Ken-Ichi Nonomura
Plants (2023) 12, 1936 DOI:10.3390/plants12101936
Meiosis is a special cell division that halves the chromosome number in preparation for fertilization. In this process, the genomic information of the parents is shuffled to generate a new gene combination (meiotic recombination), which is inherited to the next generation. The meiotic recombination machinery is widely conserved in eukaryotes and has been extensively studied. On the other hand, the mechanisms by which germ cells transition to meiosis are diverse among species, and little is known particularly in plants.
In this review, we mainly focused on meiosis of flowering plants, and explained how the archesporial cells, which produce meiocytes in the future, develop and eventually achieve meiosis with the latest findings. We also highlight the findings of our research group that the callose polysaccharide, a special plant cell wall components, plays an essential role in regulating the proper initiation timing and progression of male meiosis.
The image using the figures in our review paper was selected for the cover story of Plants Vol. 12, No. 10, including 143 articles.
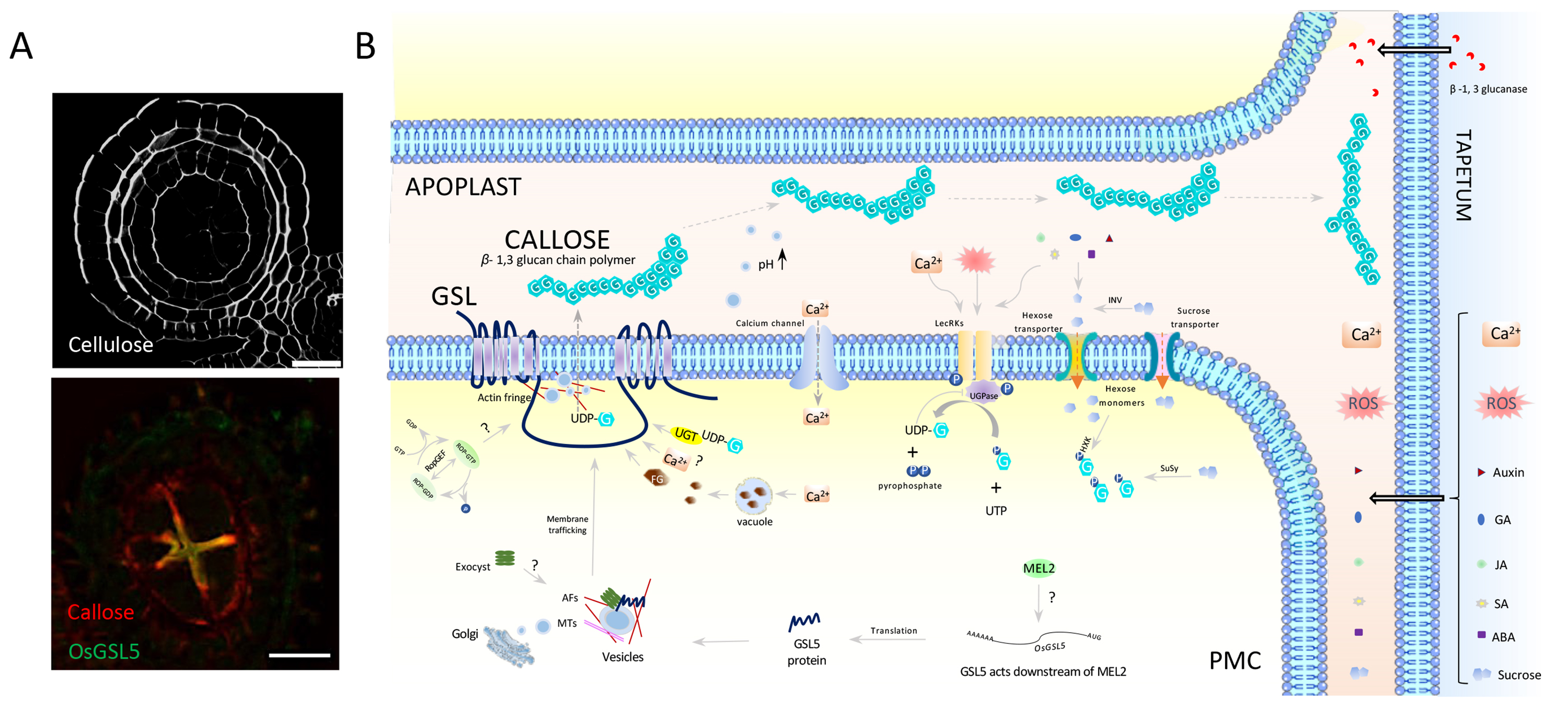
Figure: A model of callose synthesis and transport in anthers
(A) Cross sections of the anther just before meiosis. (Upper) Staining of cellulosic cell walls. (Bottom) Double immuno-staining of callose (red) and GSL5 (yellow). Bar=20µm. (B) The plasma membrane-anchored callose synthase (GSL) of male meiotic cells (PMCs) releases synthesized callose to apoplasts between PMCs (APOPLAST) just prior to meiosis transition. Callose migrates to the apoplast between PMCs and tapetum cells (TAPETUM) where GSL is absent, and eventually envelop all PMCs within the anther locule.
What is the significance of efficient assembly of the hexameric MCM2–7 helicase for DNA replication?
–A new hypothesis explaining the licensing checkpoint–
The assembly of the MCM2–7 hetero-hexamer and its significance in DNA replication.
Yuki Hatoyama and Masato T. Kanemaki.
Biochemical Society Transactions 2023 May 5 DOI:10.1042/BST20221465
To proliferate, cells must replicate chromosomal DNA, which encodes the genetic information. MCM2–7, the replication helicase that unwinds a double-strand DNA to generate a single-strand DNA, is essential for chromosomal DNA replication. The replicative MCM2–7 helicase forms a ring-shaped hexamer.
This review paper was published in Biochemical Society Transactions, a review journal published by the Biochemical Society. The review discusses how the MCM2–7 hexamer is efficiently assembled in cells and its physiological significance based on our recent report. In this review, we proposed a model in which MCMBP functions as a chaperone to promote the assembly of the MCM2–7 hexamer, resulting in the supply of sufficient amounts of MCM2–7 before DNA replication (Figure).
The existence of the licensing checkpoint was proposed, whereby the cell senses a lack of chromatin-bound MCM2–7 and arrests the cell cycle in the G1 phase. In this review, we proposed a new hypothesis that cells do not sense the amount of MCM2–7 directly. Instead, the cells with reduced MCM2–7 expression have DNA damage during DNA replication in the previous cell cycle, and they arrest the cell cycle by detecting this DNA damage in the next G1 phase.
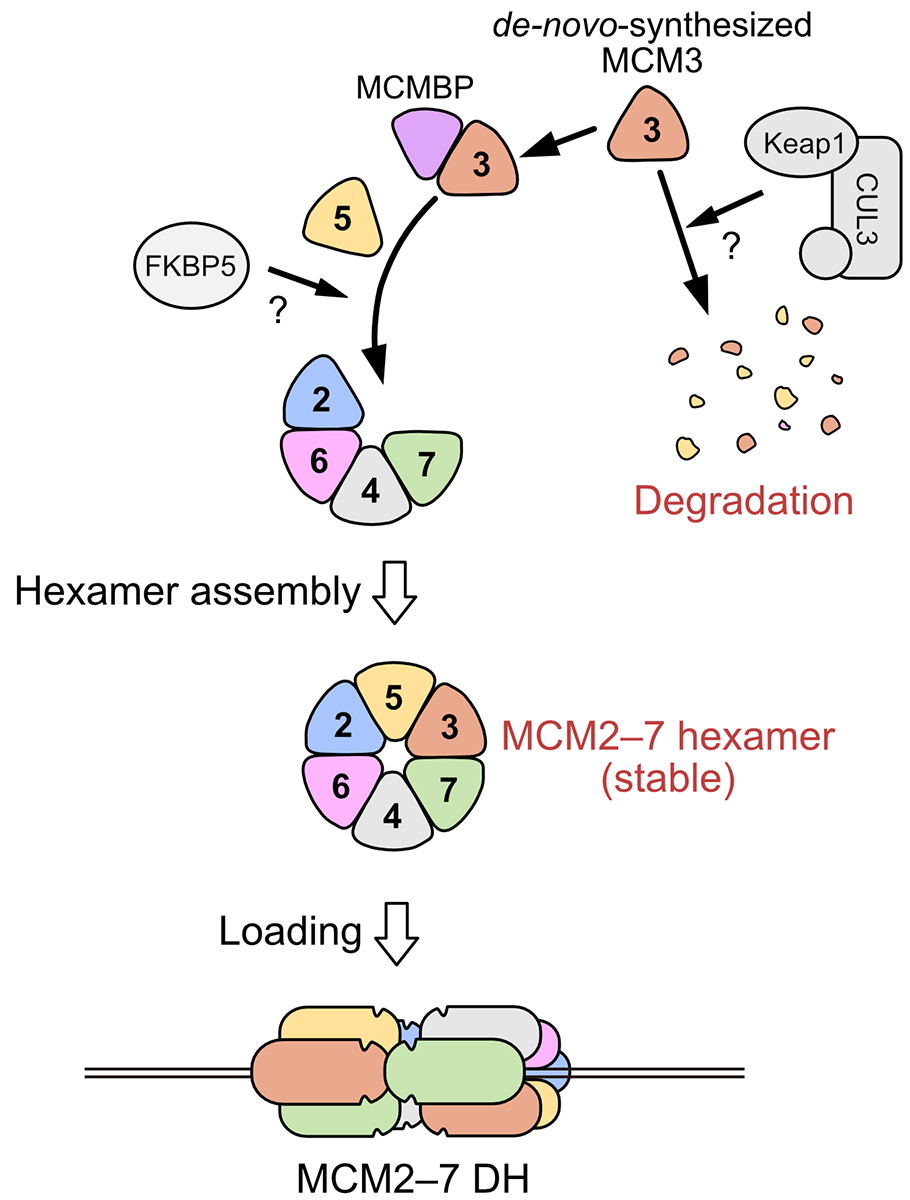
Figure: A schematic diagram of the assembling the MCM2–7 hexameric complex.
A small RNA system is involved in homology recognition between meiotic chromosomes
Press release
A small RNA system ensures accurate homologous pairing and unpaired silencing of meiotic chromosomes
Hiroaki Tabara*, Shohei Mitani, Megumi Mochizuki, Yuji Kohara and Kyosuke Nagata *: corresponding author
The EMBO Journal 2023 April 20 DOI:10.15252/embj.2020105002
Meiosis is a specialized mode of cell division that produces haploid gametes. During meiosis, chromosomes with homologous partners undergo synaptonemal complex (SC)-mediated pairing, while the remaining unpaired regions such as most parts of the sex chromosomes in males are heterochromatinized through unpaired silencing.
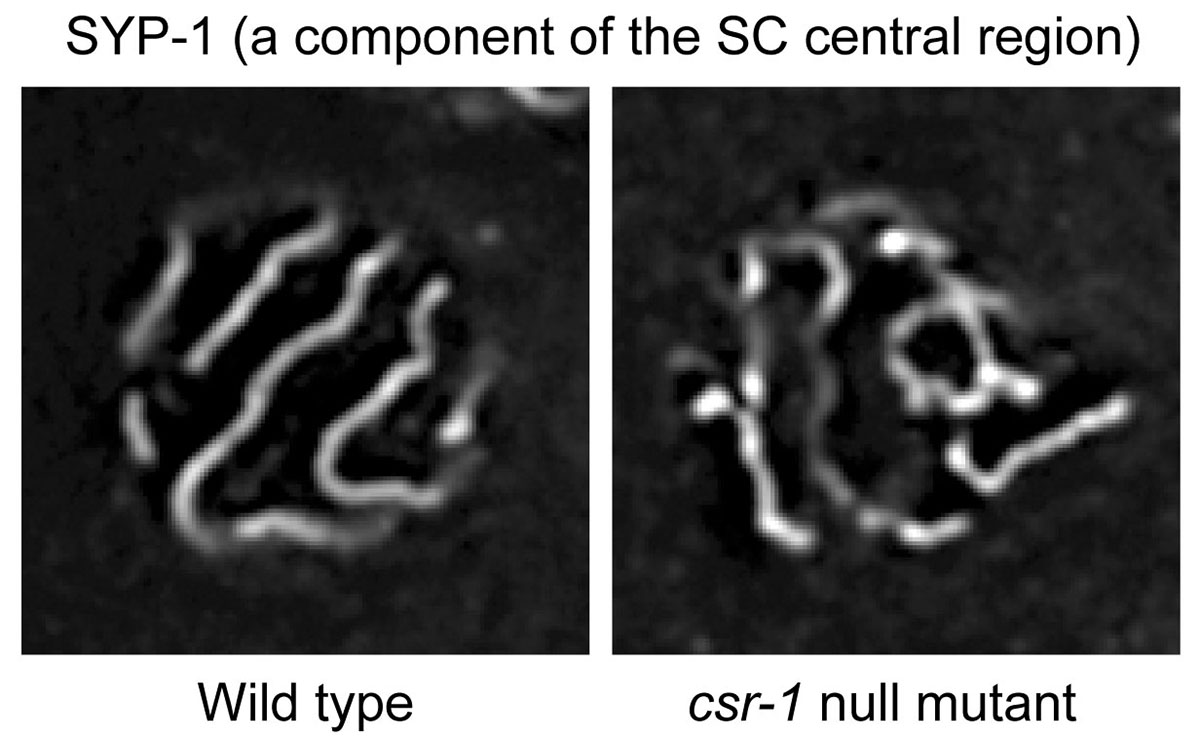
A study, published in The EMBO Journal, introduces that a small RNA system regulates meiotic pairing of homologous chromosomes in the nematode Caenorhabditis elegans. Homolog pairing initiation depends on species-specific mechanisms involving interactions between pericentric heterochromatin, centromeres, telomeres, and/or pairing center sequences. Subsequently, the SC establishes the pairing of other regions of the homologous chromosomes. In plants and mammals, DNA double-strand breaks (DSBs) are important or partly important for synapse formation between homologous chromosomes. Mysteriously, in C. elegans and fruit fly, DSBs are not necessary for synapse formation, implying the presence of an unclarified mechanism for recognizing homologous chromosomes during SC formation.
RNA interference (RNAi) is a form of RNA-mediated sequence-specific regulation of gene expression in eukaryotes. Small RNAs and Argonaute proteins play key roles in the mechanism of RNAi. Researchers at National Institute of Genetics, Tokyo Women’s Medical University, and University of Tsukuba, in Japan, found new relationships between SC formation on paired chromosomes, unpaired chromosome heterochromatinization, and an Argonaute–small RNA pathway (endogenous RNAi). The C. elegans Argonaute proteins, CSR-1 and its paralog interacting with small RNAs, are required for SC formation between chromosome pairs with accurate homology and are also involved in the condensation of unpaired chromosomes. The SC is composed of layers of strings, with two lateral elements sandwiching a central region. CSR-1 in nuclei and meiotic cohesin, constituting the SC lateral elements, were associated with non-simple DNA repeats expressing small RNAs and long non-coding RNAs (lncRNAs), and also associated weakly with coding genes. This study proposes a zipper model, in which the accurate homolog pairing is promoted via the inter-chromosomal recognition of chromosome-associated lncRNAs by the Argonaute-mediated small RNA system. The meiotic difference between paired or unpaired chromosomes is also described.
Reference
Tabara H, Mitani S, Mochizuki M, Kohara Y, Nagata K (2023) A small RNA system ensures accurate homologous pairing and unpaired silencing of meiotic chromosomes. EMBO J, in press. DOI: 10.15252/embj.2020105002.
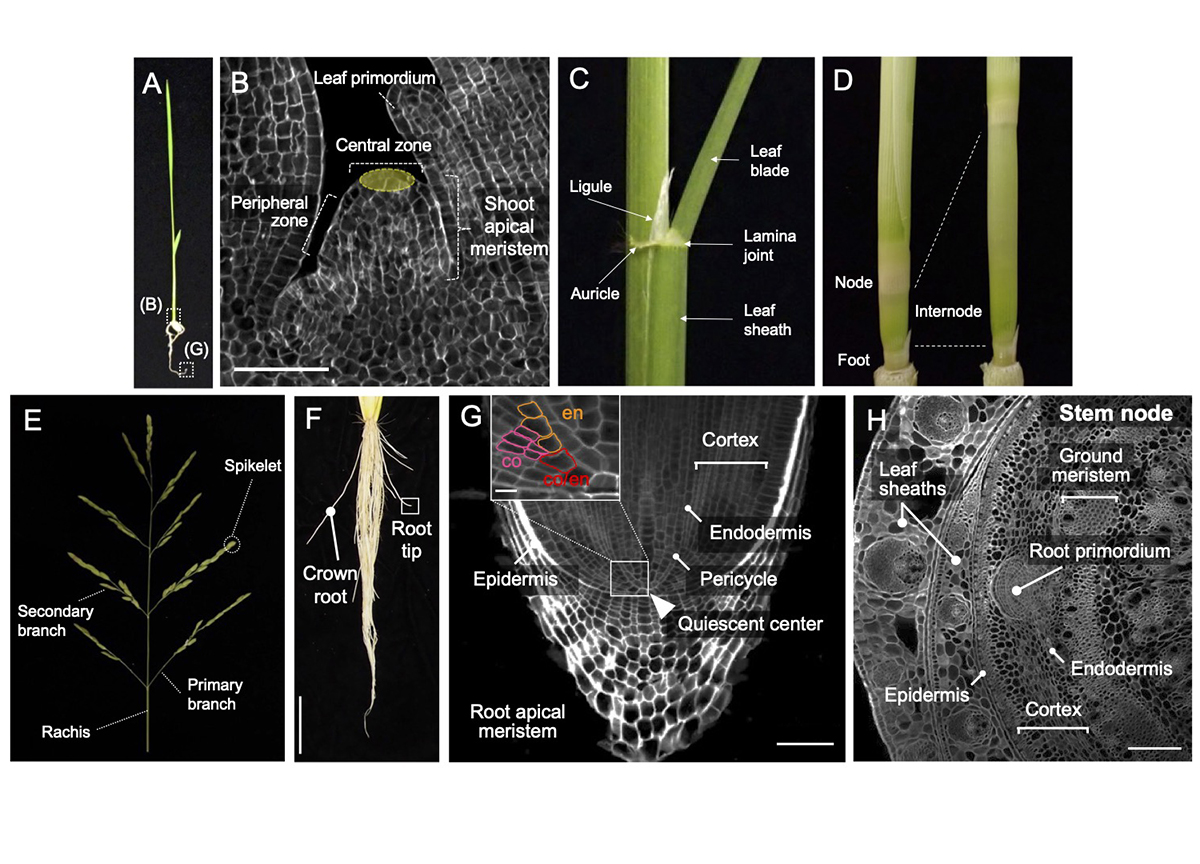
Genetic basis controlling rice plant architecture and its modification for breeding
Genetic basis controlling rice plant architecture and its modification for breeding
Wakana Tanaka, Takaki Yamauchi, *Katsutoshi Tsuda *Corresponding author
Breeding Science (2023) 73, 3-45 DOI:10.1270/jsbbs.22088
Rice (Oryza sativa) is derived from the ancestral wild species Oryza rufipogon, which began to be cultivated about 10,000 years ago. In this process, the morphology of rice has undergone significant changes through human selection. For example, cultivated rice has a more upright stature for dense planting, generates shorter stems to prevent lodging (the bending over of the stem to the ground), and produces more grains in inflorescences compared to its ancestor. Recent molecular genetics has identified numerous important regulators and molecular mechanisms in development. Importantly, many regulators have been utilized as weak alleles in the process of domestication and subsequent breeding. This review aimed to comprehensively summarize a wide range of knowledge in rice developmental biology for each organ, including leaf, stem, inflorescence, and root, and to highlight examples utilized in breeding. This review will help to learn the current understanding of rice morphogenesis and provide a theoretical foundation for future crop improvement.
This work was supported by JSPS Grants (22K06267, 22H02308, 22H02319).
Euchromatin forms condensed chromatin domain.
Press release
Condensed but liquid-like domain organization of active chromatin regions in living human cells
Tadasu Nozaki*, Soya Shinkai*, Satoru Ide*, Koichi Higashi*, Sachiko Tamura, Masa A. Shimazoe, Masaki Nakagawa, Yutaka Suzuki, Yasushi Okada, Masaki Sasai, Shuichi Onami, Ken Kurokawa, Shiori Iida, Kazuhiro Maeshima *co-first authors
Science Advances (2023) 9, eadf1488 DOI:10.1126/sciadv.adf1488
![]() Press release (In Japanese only)
Press release (In Japanese only)
The human genome chromatin can be classified into euchromatin and heterochromatin, which have high and low transcription activities, respectively. In the classical view, it was believed that euchromatin has an open and decondensed structure, while heterochromatin is highly condensed.
A research team led by Professor Kazuhiro Maeshima of Genome Dynamics Laboratory (NIG), including a graduate student (currently a Research Associate at Harvard Univ.) Tadasu Nozaki, Assistant Professor Satoru Ide, Technical Stuff Sachiko Tamura, SOKENDAI graduate student Masa A. Shimazoe, SOKENDAI graduate student (JSPS Research Fellow DC2) Shiori Iida, together with Assistant Professor Koichi Higashi and Professor Ken Kurokawa of Genome Evolution Laboratory (NIG), and Senior Research Scientist Soya Shinkai and Team Leader Shuichi Onami of RIKEN BDR, have found euchromatin forms condensed chromatin domains. This finding suggests an innovative model that condensed structure is the default state of chromatin.
In this work, motions of fluorescently labeled nucleosomes in euchromatin in living human cells were investigated with a super-resolution microscope. Two neighboring nucleosomes were observed simultaneously with different colored fluorescent dyes. The research team discovered that movements of nucleosome pairs within 150 nm in the distance were correlated. This shows that euchromatin forms irregularly condensed chromatin domains with ~150 nm diameters. Further analysis revealed that nucleosomes within a domain behave like a liquid. On the other hand, euchromatin is stable at the chromosome scale, and exhibits a solid-like behavior.
This liquid-like behavior of nucleosomes can facilitate transcription, DNA replication, and repair in condensed domains. Also, the condensed domains can protect DNA from radiation damage. Solid-like behavior of chromatin at the chromosome scale can contribute to maintaining the genome by preventing entanglements or breaks of long chromatin. Chromatin is locally dynamic and reactive, and globally stable.
This work was supported by JSPS Fellowship, JSPD grants (21H02453, 22H05606, 21H02535, 20H05550, 20H05936, 16H06279(PAGS), 22H04925 (PAGS)), a Japan Science and Technology Agency CREST grant (JPMJCR15G2), JST SPRING(JPMJSP2104), and the Uehara Memorial Foundation.
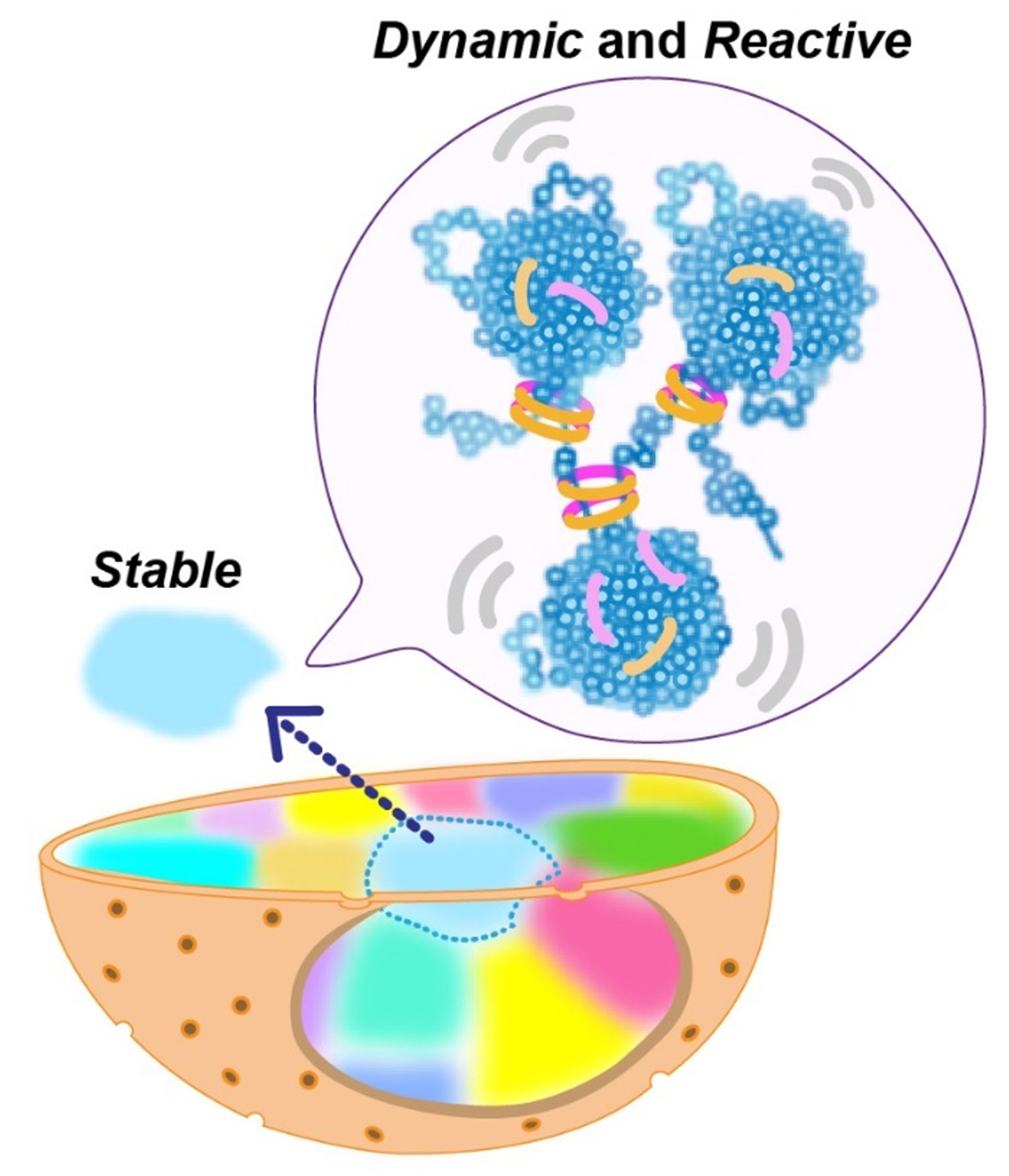
Figure: Nucleosomes form irregularly condensed chromatin domains by their intrinsic sticky nature (blue clusters in the balloon). Nucleosomes show dynamic liquid-like behavior inside a domain, which promotes DNA transactions. On the other hand, the solid-like behavior of chromatin on the chromosome scale helps to maintain the genome by preventing entanglements or breaks of long chromatin.
Video: Video of nucleosome pairs at euchromatin labeled by two different fluorescent dyes and recorded simultaneously. The upper row shows a neighboring nucleosome pair with correlated movements. The lower row depicts a distant nucleosome pair with uncorrelated movements.
The RAD51 recombinase promotes the reversed-fork formation behind the CMG helicase
Kanemaki Group / Molecular Cell Engineering Laboratory
RAD51 bypasses the CMG helicase to promoter fork reversal.
Liu W, Saito Y, Jackson J, Bhowmick R, Kanemaki MT, Vindigni A and *Cortez D.
Science (2023) 380, 382-387 DOI:10.1126/science.add7328
Replication forks are known to stall due to replication stress, and electron microscopy observation has revealed the occurrence of the fork reversal structure, also known as the chicken-foot structure. It was previously known that the RAD51 recombinase is required for the formation of the reversed forks. However, it was unclear whether CMG helicase, which was responsible for moving the replication fork, remained present in the reversed forks and, if so, where it was located. This study analyzed RAD51 mutants and found that the reversed fork structure forms behind the CMG helicase and that the requirement for RAD51 in fork reversal formation disappears when CMG helicase is absent.
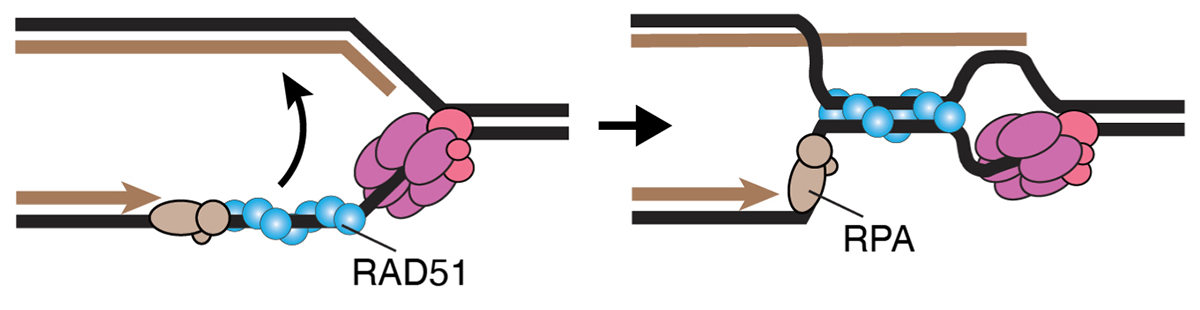
This study was conducted as collaborative research between Professor David Cortez at Vanderbilt University and the Kanemaki Laboratory at NIG. The Kanemaki Laboratory generated AID2 cells that can degrade MCM proteins, which is a main component of the CMG helicase.
New assistant professor joins NIG
New assistant professor joins NIG as of April 1, 2023.
KUROKAWA, Yumiko : Murayama Group • Chromosome Biochemistry Laboratory
New laboratories established in the Center for Frontier Research
Two Associate professors have joined Center for Frontier Research as of April 1, 2023.
associate professor
▶SASAKI, Mariko:Center for Frontier Research, Laboratory of Gene Quantity Biology
▶YAMAMICHI, Masato:Center for Frontier Research, Theoretical Ecology and Evolution Laboratory
Center for Frontier Research is an incubation center to simultaneously develop two elements; human resources and new research fields. Promising young scientists conduct research as principal investigator (tenure-track associate professor) to explore new frontiers in genetics and related areas, taking advantage of NIG’s research infrastructure and various support systems.
Ms. Piu Banerjee (Laboratory of Mammalian Neural Circuits) received the Dean’s Award

Right: Prof. Iwasato (Dean/Supervisor)
The Dean’s Award is given to students who complete the PhD program with research activities worthy of special recognition at each school of the Graduate University for Advanced Studies, SOKENDAI.
Ms. Piu Banerjee (Laboratory of Mammalian Neural Circuits, Iwasato Laboratory) received the School of Life Science Dean’s Award for the second semester of the 2022 academic year.
Piu Banerjee (Laboratory of Mammalian Neural Circuits, Iwasato Laboratory)
Thesis Title: Study of Spontaneous Activity in the Peripheral Structure of Developing Mouse Somatosensory System
Ms. Piu Banerjee also received the Morishima Award from the Department of Genetics.

















