Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Feeding on phototoxic algae: training for unicellular predators to evolve to algae
Responses of unicellular predators to cope with the phototoxicity of photosynthetic prey
Akihiro Uzuka*, Yusuke Kobayashi*, Ryo Onuma*, Shunsuke Hirooka, Yu Kanesaki, Hirofumi Yoshikawa, Takayuki Fujiwara, and Shin-ya Miyagishima
(*Co-first authors)
Nature Communications 10, 5606 (2019) DOI:10.1038/s41467-019-13568-6
Link to “Behind the paper” in Nature Ecology and Evolution.
Feeding on unicellular photosynthetic organisms by unicellular eukaryotes is the base of the aquatic food chain and evolutionarily led to the establishment of photosynthetic endosymbionts/organelles. Photosynthesis generates reactive oxygen species and damages cells; thus, photosynthetic organisms possess several mechanisms to cope with the stress. Here, we demonstrate that photosynthetic prey also exposes unicellular amoebozoan and excavate predators to photosynthetic oxidative stress. Upon illumination, there is a commonality in transcriptomic changes among evolutionarily distant organisms feeding on photosynthetic prey. One of the genes commonly upregulated is a horizontally transferred homolog of algal and plant genes for chlorophyll degradation/detoxification. In addition, the predators reduce their phagocytic uptake while accelerating digestion of photosynthetic prey upon illumination, reducing the number of photosynthetic cells inside the predator cells, as this also occurs in facultative endosymbiotic associations upon certain stresses. Thus, some mechanisms in predators observed here probably have been necessary for evolution of endosymbiotic associations.
This study was supported by JSPS KAKENHI to S.-y.M. (16K14791 and 17H01446) and to A.U. (17J08575), and by the MEXT Program for Strategic Research Foundation at Private Universities to Yu.K. and H.Y. (2013–2017 S1311017).
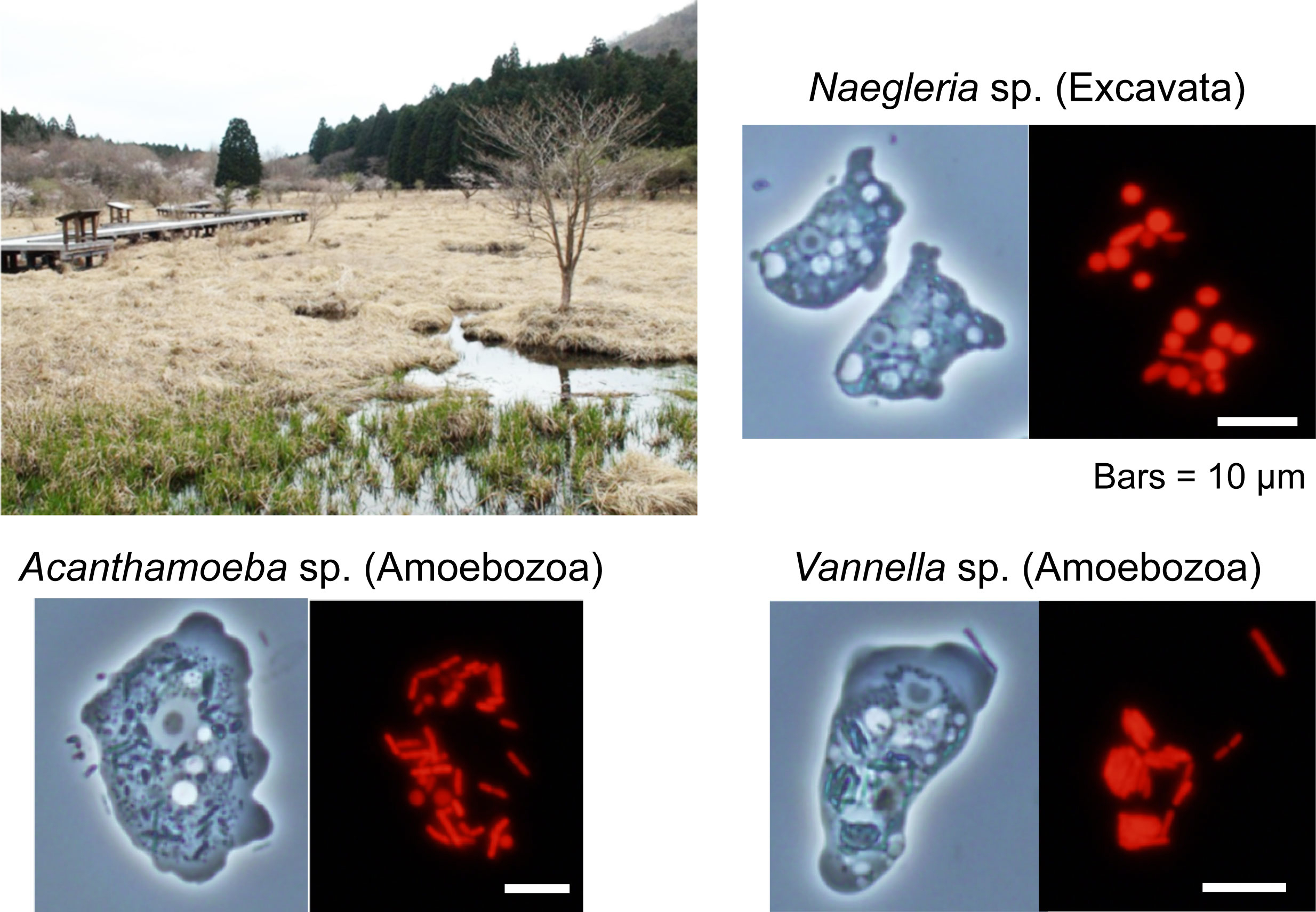
Figure: Three species of amoebae feeding on algae were isolated from Kodanuki Marsh in Fujinomiya, Japan on April 23, 2013. Although they belong to different eukaryotic supergroups (two belong to Amoebozoa, one to Excavata), they have independently evolved a similar strategy to cope with the photosynthetic oxidative stress that caused by the ingested photosynthetic prey. Red is fluorescence of chlorophyll (a photosynthetic pigment).
Canceled: The NIG International Symposium on Molecular Mechanism of Chromosome Replication
Repulsive signal contributes to proper termination of neuronal migration
Semaphorin 6A–Plexin A2/A4 interactions with radial glia regulate migration termination of superficial layer cortical neurons.
Yumiko Hatanaka, Takahiro Kawasaki, Takaya Abe, Go Shioi, Takao Kohno, Mitsuharu Hattori, Akira Sakakibara, Yasuo Kawaguchi & Tatsumi Hirata
iScience 21, pp 359-374, 2019 DOI:10.1016/j.isci.2019.10.034
Precise regulation of neuronal migration termination is crucial for the establishment of brain cytoarchitectures. However, little is known how neurons terminate migration. We found that superficial layer cortical neurons (SLNs) migrate beyond their final destination and ectopically invade layer 1 in Plexin (Plxn) A2/A4 double-knockout mice as well as Semaphorin (Sema) 6A knockout mice. Cell-targeted gene expression and conditional knockouts indicated that Sema6A on radial glial cells and PlxnA2/A4 on SLNs are involved in this process. Given that Sema6A–PlxnA2/A4 trans-interactions generally induce a repulsive reaction, our results suggest that Sema6A–PlxnA2/A4 interaction elicits repulsion and weakens migrating neuron–substrate interactions, leading to migration termination.
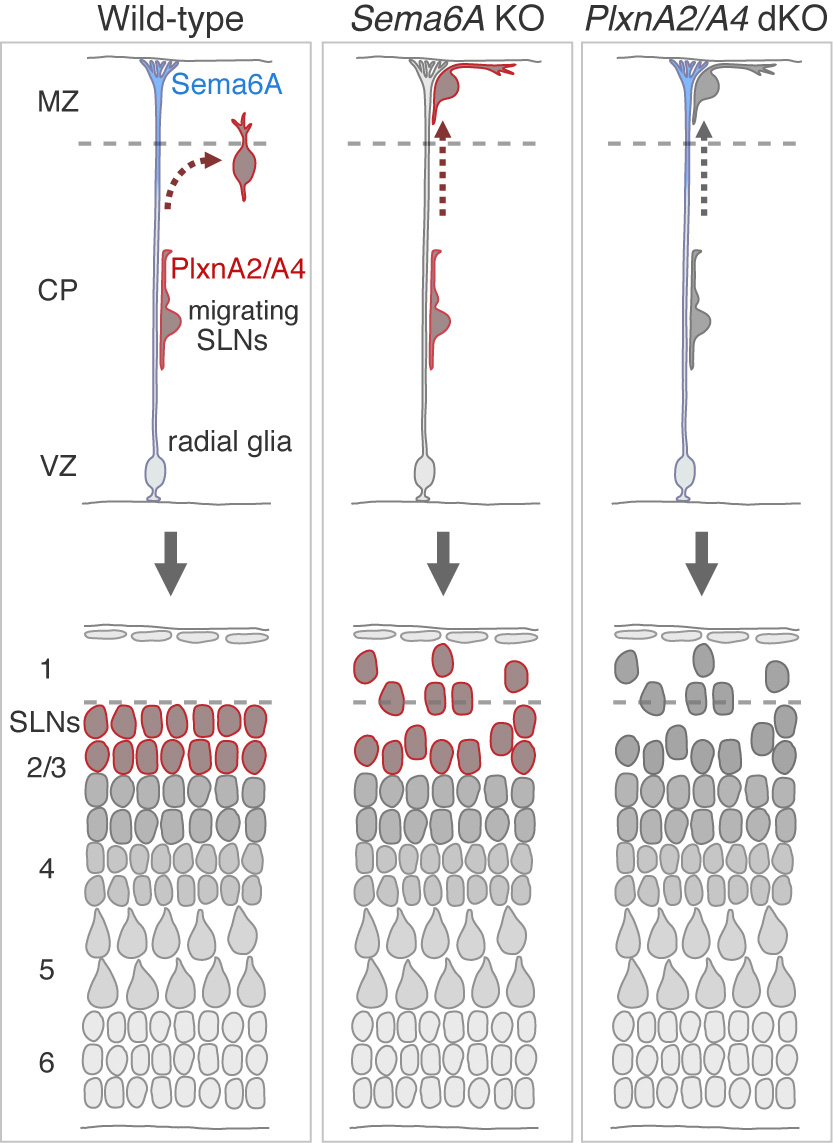
Figure: Cortical neurons migrate toward the pial surface along radial glial cell fibers. Sema6A on radial glial cells and PlxnA2/A4 on migrating neurons are necessary for proper migration termination of superficial layer cortical neurons.
A novel katanin-tethering machinery accelerates cytokinesis
Press release
A novel katanin-tethering machinery accelerates cytokinesis
Takema Sasaki, Motosuke Tsutsumi, Kohei Otomo, Takashi Murata, Noriyoshi Yagi, Masayoshi Nakamura, Tomomi Nemoto, Mitsuyasu Hasebe, Yoshihisa Oda
Current Biology 29, 1-11 DOI:10.1016/j.cub.2019.09.049
Press release (In Japanese only)
Cytokinesis is fundamental for cell proliferation. In plants, a bipolar short-microtubule array forms the phragmoplast, which mediates vesicle transport to the midzone and guides the formation of cell walls that separate the mother cell into two daughter cells. The phragmoplast centrifugally expands toward the cell cortex to guide cell-plate formation at the cortical division site. Several proteins in the phragmoplast midzone facilitate the anti-parallel bundling of microtubules and vesicle accumulation. However, the mechanisms by which short microtubules are maintained during phragmoplast development, in particular, the behavior of microtubules at the distal zone of phragmoplasts, are poorly understood. Here, we show that a plant-specific protein, CORTICAL MICROTUBULE DISORDERING 4 (CORD4), tethers the conserved microtubule-severing protein katanin to facilitate formation of the short-microtubule array in phragmoplasts. CORD4 was specifically expressed during mitosis and localized to preprophase bands and phragmoplast microtubules. Custom-made two-photon spinning disk confocal microscopy revealed that CORD4 rapidly localized to microtubules in the distal phragmoplast zone during phragmoplast assembly at late anaphase and persisted throughout phragmoplast expansion. Loss of CORD4 caused abnormally long and oblique phragmoplast microtubules and slow expansion of phragmoplasts. The p60 katanin subunit, KTN1, localized to the distal phragmoplast zone in a CORD4-dependent manner. These results suggest that CORD4 tethers KTN1 at phragmoplasts to modulate microtubule length, thereby accelerating phragmoplast growth. This reveals the presence of a distinct machinery to accelerate cytokinesis by regulating the action of katanin.
Source: Sasaki et al., (2019) Current Biology 29, 1-11, DOI: 10.1016/j.cub.2019.09.049
Summer Internship at NIG “NIGINTERN2020”
Microfocus X-ray CT (microCT) Imaging of Actinia equina (Cnidaria), Harmothoe sp. (Annelida), and Xenoturbella japonica (Xenacoelomorpha).
Microfocus X-ray CT (microCT) Imaging of Actinia equina (Cnidaria), Harmothoe sp. (Annelida), and Xenoturbella japonica (Xenacoelomorpha).
Maeno A, Kohtsuka H, Takatani K, Nakano H.
Journal of Visualized Experiments 150 e59161 1-9 DOI:10.3791/59161
Traditionally, biologists have had to rely on destructive methods such as sectioning in order to investigate the internal structures of opaque organisms. Non-destructive microfocus X-ray computed tomography (microCT) imaging has become a powerful and emerging protocol in biology, due to technological advancements in sample staining methods and innovations in microCT hardware, processing computers, and data analysis software. However, this protocol is not commonly used, as it is in the medical and industrial fields. One of the reasons for this limited use is the lack of a simple and comprehensible manual that covers all of the necessary steps: sample collection, fixation, staining, mounting, scanning, and data analyses. Another reason is the vast diversity of metazoans, particularly marine invertebrates. Because of marine invertebrates’ diverse sizes, morphologies, and physiologies, it is crucial to adjust experimental conditions and hardware configurations at each step, depending on the sample. Here, microCT imaging methods are explained in detail using three phylogenetically diverse marine invertebrates: Actinia equina (Anthozoa, Cnidaria), Harmothoe sp. (Polychaeta, Annelida), and Xenoturbella japonica (Xenoturbellida, Xenacoelomorpha). Suggestions on performing microCT imaging on various animals are also provided.
Source: Maeno A. et al, (2019) J Vis Exp. 6 Augst, DOI:10.3791/59161.
Inquiries : Technical Specialist, MAENO, Akiteru(amaeno@nig.ac.jp)
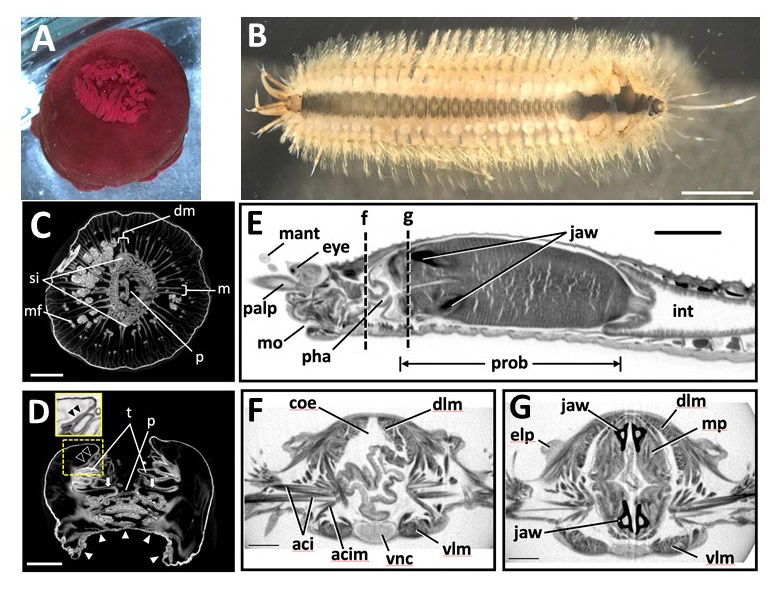
Figure: Marine invertebrate animals observed in this study. (A,B)
(A) Live anesthetized Actinia equina (Anthozoa, Cnidaria).
(B) Live anesthetized Harmothoe sp. (Polychaeta, Annelida).Most of the elytra were already missing at this stage, with only four remaining near the posterior end. Scale bars = 3 mm.
Scanned and reconstructed images of marine invertebrates. (C-G)
(C) Transverse and (D) longitudinal sections of Actinia equina. Scale bars in C, D = 3 mm.
(E-G) Harmothoe sp. (Polychaeta, Annelida). (E) Sagittal section of the anterior part. (F, G) Transverse section at the dotted lines f and g in (E). Scale bars: E = 1 mm; F, G = 0.3 mm.
Volume rendering image and transverse section movie of the whole body of Harmothoe sp. 6 sec to 16 sec: 3D volume rendering image. Top: left view, bottom: frontal view. 17 sec to 1min 42 sec: Transverse section movie. The position of the section is shown with a moving green line on the sagittal section image at top. From 17 sec to 53 sec, transverse section movie of the anterior part of the specimen generated from Pinpoint Scan is shown at the left bottom. From 1 min 7 sec to 1 min 14 sec, a 3D model made using Imaris software showing the positions of major organs is present at right top. Anatomical supervision: Masaatsu Tanaka (Kagoshima University)
This technique is one of the bases of following researches.
・ Evolution of Shh endoderm enhancers during morphological transition from ventral lungs to dorsal gas bladder
・ Regulation of internode patterning and vein anastomosis in maize stems.
・ Combination of multiple Shh enhancers controls tooth development in mouse
・Development of organs in living whole embryo/larval grafts in zebrafish
・ Enhancer adoption changes limb morphology
・When do male and female differences appear in the development of beetle horns? (National Institute for Basic Biology )
・A mouse model of human chromosomal disorder, partial trisomy distal 4q
A novel central olfactory circuit revealed by a newly developed neuronal birthdate tagging method
A Novel Birthdate-Labeling Method Reveals Segregated Parallel Projections of Mitral and External Tufted Cells in the Main Olfactory System
Tatsumi Hirata, Go Shioi, Takaya Abe, Hiroshi Kiyonari, Shigeki Kato, Kazuto Kobayashi, Kensaku Mori and Takahiko Kawasaki
eNeuro 31, ENEURO.0234-19.2019, 2019 DOI:10.1523/ENEURO.0234-19.2019
Odorant receptors form an ordered odorant map on the main olfactory bulb. This spatial representation disappears in most subsequent targets by “diffuse divergence and random convergence” of olfactory bulb projections. We revisited these projections using a newly developed method that can genetically dissect distinct subsets of olfactory bulb projection neurons based on their neuronal birthdates. Our birthdate tag analysis exposed parallel segregated projections formed by early-born mitral and late-born external tufted cells in otherwise apparently random olfactory networks. The results suggest that these parallel pathways extract unique features of information from the common olfactory input and process these features in a way similar to “color”, “orientation” or “direction” in the visual system. Importantly, the birthdate tag method can pave the way for deciphering the functional meaning of these individual pathways in the future.
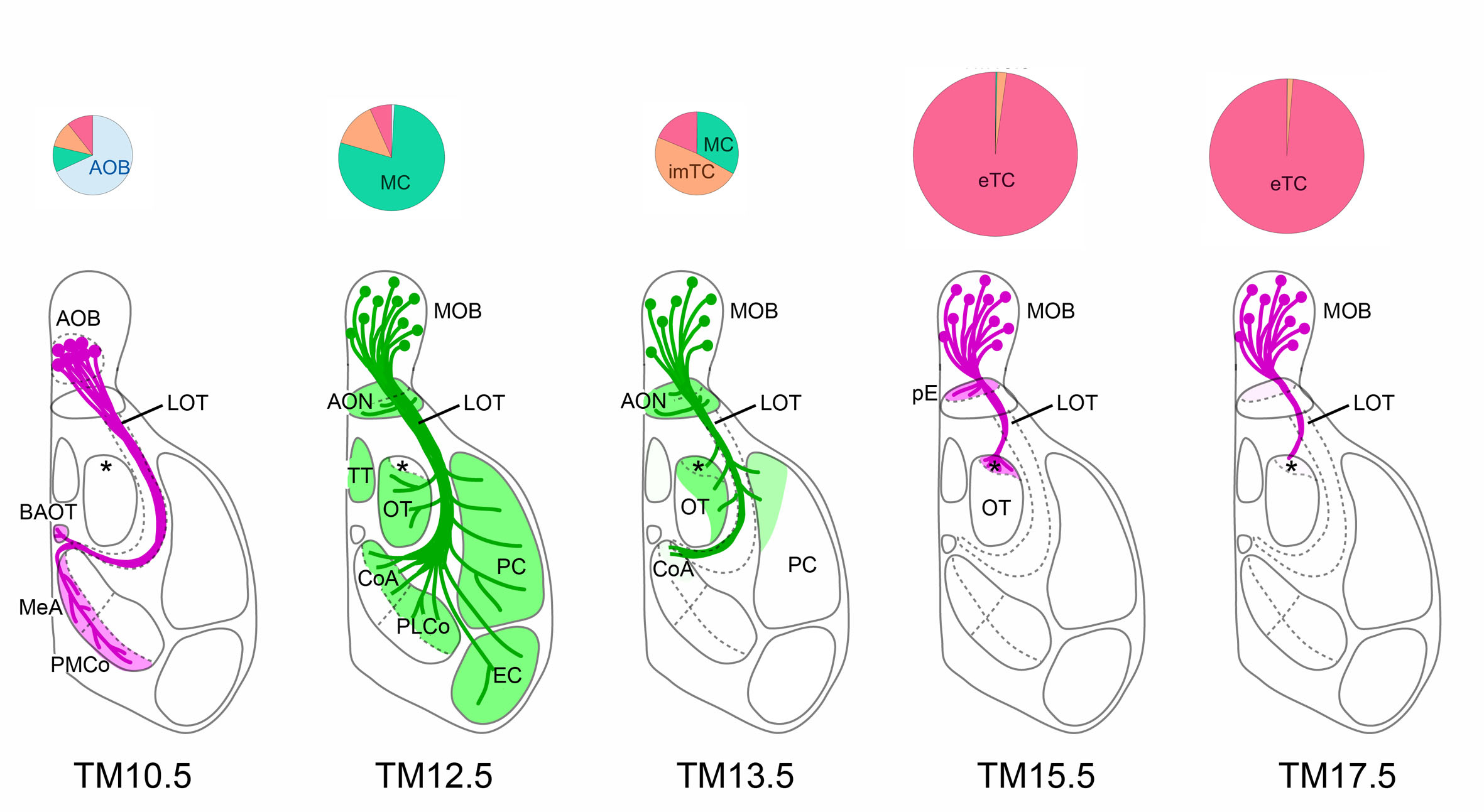
Figure: Dissection of olfactory bulb projection neurons and their axon trajectories using neuronal birthdate tagging
Depending on tamoxifen injection stages (TM10.5~17.5), different classes of neurons such as accessory olfactory bulb neurons (AOB), mitral cells (MC) or tufted cells (TC) are tagged (pie charts), and their axon trajectories are revealed (bottom diagrams).
- This method is one of the basises of“NeuroGT Database”, a brain atlas ofneurogenic tagging CreER drivers”(NeuroGT).
Identification of RNA-binding proteins involved in the formation and maintenance of egg precursors
ELAVL2-directed RNA regulatory network drives the formation of quiescent primordial follicles
Yuzuru Kato, Tokuko Iwamori, Yoichirou Ninomiya, Takashi Kohda, Jyunko Miyashita, Mikiko Sato, and Yumiko Saga
EMBO reports e48251, 2019 DOI:10.15252/embr.201948251
Long‐term reproduction of mammalian females is achieved by controlled sustainability and growth of the most primitive ovarian follicles called primordial follicles, which are composed of a single oocyte and a few of the supporting somatic cells, granulosa cells. Given the importance of primordial follicles as a finite reservoir of eggs in mammalian ovaries, understanding the molecular mechanisms underlying the formation and sustainability of primordial follicles is essential for reproductive biology and medicine.
Kato et al., demonstrated that RNA-binding proteins ELAVL2 and DDX6 drive the assembly of P-body-like cytoplasmic RNP granules in oocytes and the formation of primordial follicles collaborating with Kyushu university and Yamanashi University. The research group revealed that an RNA-binding protein ELAVL2 is indispensable for the formation of primordial follicles in mice. Subsequently, the group identified mRNAs that interact with ELAVL2, among which they found that mRNAs encoding components of cytoplasmic RNP granules such as P-bodies were enriched. Furthermore, deletion of Ddx6, one of the targets of ELAVL2 and a gene encoding crucial component of P-bodies, exhibited disassembly of P-body-like granules in oocytes and defective formation of primordial follicles. These findings revealed that the ELAVL2-directed post-transcriptional network is crucial for primordial follicle formation.
This study was supported by Grant-in-Aid for Scientific Research (C) to Y.K. (No.17K07422) and (A) to Y.S. (No.26251025) from JSPS and by Grant-in-Aid for Scientific Research on Innovative Area to Y.K. (No.26114512, 16H01259, 19H05246) and T.I. (No.26114506) from MEXT.
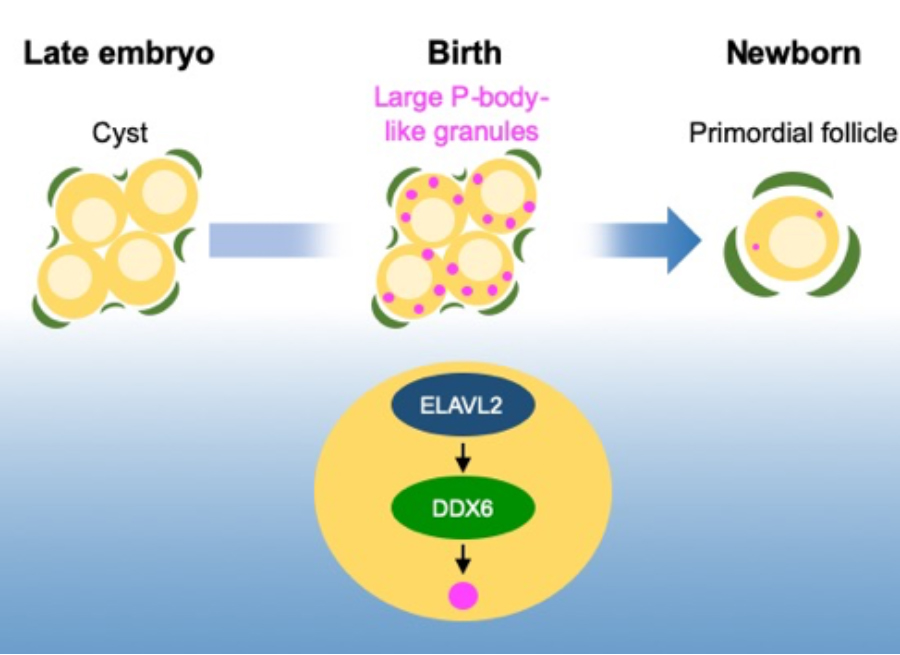
Figure: Primordial follicles arise from oocyte in cyst shortly after birth. In oocytes, large P-body-like cytoplsamic RNP granules are formed depending on the function of RNA-binding proteins ELAVL2 and DDX6. Deletion of Elavl2 or Ddx6 results in the defective assembly of the granules and the formation of primordial follicles in mice.
Prof. Maeshima was interviewed by JCB
Prof. Kazuhiro Maeshima at Genome Dynamics Laboratory was interviewed by the Journal of Cell Biology (Rockefeller University Press) on his career, mind, and work, and the interview article has been online in People & Ideas section of JCB.
▶ Kazuhiro Maeshima: Excitement under the microscope
JCB People & Ideas are interviews intended to tell the story of individual scientists and how they got to where they are today. The section aims to provide readers with a sense not only of their scientific journey but also their personal journey, highlighting unique experiences or interesting details about them and their work.
Recovered: About the error that seminar information is not displayed correctly
The problem that seminar information was not displayed correctly in some browser versions has been fixed.
Thank you for your understanding.
Fluid-like behavior of chromatin in living human cell
Press release
Organization of fast and slow chromatin revealed by single-nucleosome dynamics
S. S. Ashwin, Tadasu Nozaki, Kazuhiro Maeshima, and Masaki Sasai
PNAS first published September 16, 2019 DOI:10.1073/pnas.1907342116
Press release (In Japanese only)
Understanding chromatin organization and dynamics is important, since they crucially affect DNA functions. In this study, we investigate chromatin dynamics by statistically analyzing single-nucleosome movement in living human cells. Bimodal nature of the mean square displacement distribution of nucleosomes allows for a natural categorization of the nucleosomes as fast and slow. Analyses of the nucleosome–nucleosome correlation functions within these categories along with the density of vibrational modes show that the nucleosomes form dynamically correlated fluid regions (i.e., dynamic domains of fast and slow nucleosomes). Perturbed nucleosome dynamics by global histone acetylation or cohesin inactivation indicate that nucleosome–nucleosome interactions along with tethering of chromatin chains organize nucleosomes into fast and slow dynamic domains. A simple polymer model is introduced, which shows the consistency of this dynamic domain picture. Statistical analyses of single-nucleosome movement provide rich information on how chromatin is dynamically organized in a fluid manner in living cells.
This research was supported by JST CREST(JPMJCR15G2), JSPS Kakenhi (JP19H05258, JP19H05273, JP19H01860, JP16H04746) and Takeda Science Foundation, RIKEN Pioneering Project、NIG Joint (2016-A2(6))
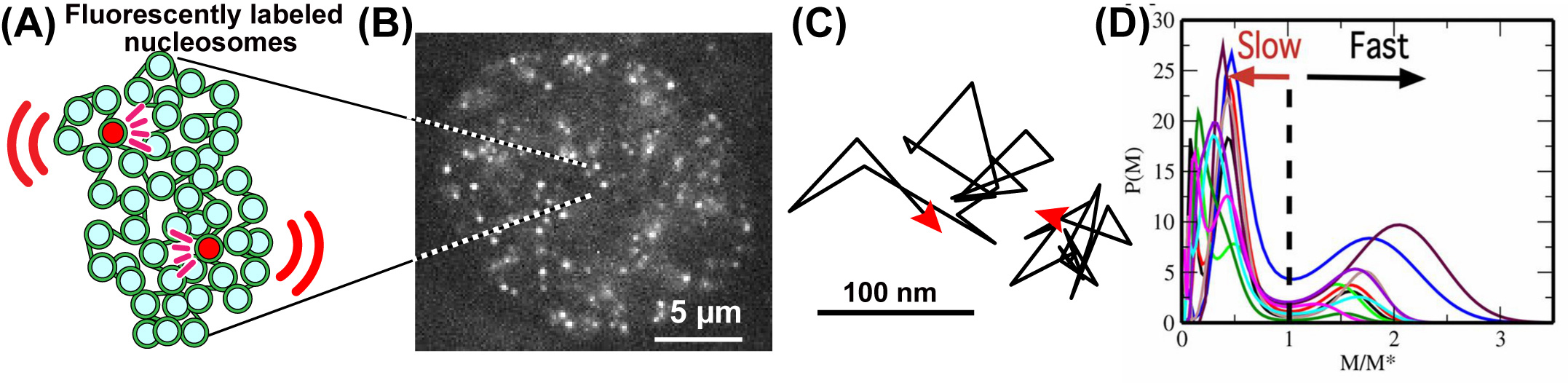
Fig: (A) A small fraction of nucleosomes, where DNA is wrapped around histone proteins, was fluorescently labeled (red). The labeled nucleosome movements can be tracked at super-resolution. (B) A single-nucleosome image of a living HeLa cell. (C) Representative two trajectories of the tracked single nucleosomes. (D) The distribution of MSD of single nucleosomes is plotted for 10-cell samples as functions of M/M*, where M* is M at the minimum between 2 peaks of the distribution.
Video: Raw video of single nucleosomes in the living HeLa cell. From Nozaki et al., (2017) Molecular Cell.
A new homologous recombination factor, HROB, controls the MCM8–MCM9 pathway
Control of homologous recombination by the HROB–MCM8–MCM9 pathway
Nicole Hustedt, Yuichiro Saito, Michal Zimmermann, Alejandro Álvarez-Quilón, Dheva Setiaputra, Salomé Adam, Andrea McEwan, Jing Yi Yuan, Michele Olivieri, Yichao Zhao, Masato T. Kanemaki, Andrea Jurisicova, and Daniel Durocher
Genes & Development online advanced publication DOI:10.1101/gad.329508.119
Most organisms including humans have two copies of genomic DNA, each of which is originated from the parents. Homologous Recombination (HR) is a reaction, which copy the sequence of donor DNA and paste it to the recipient DNA. HR plays a critical role in meiosis for gametogenesis and in damaged DNA repair during DNA replication or after irradiation, the latter of which is important to prevent cell death and cancer formation.
HR searches the donor sequence and synthesize DNA to copy and paste it to the recipient ones. Despite having a good understanding of the protein roles in the earlier steps in HR, much less is known about HR-associated DNA synthesis. We have studied MCM8–9, which is essential for HR-associated DNA synthesis. As a collaboration with the group led by Prof. Daniel Durocher at University of Toronto, we identified a new factor, HROB, which has an important role for loading MCM8–9 at the damaged DNA sites. Similar to the phenotypes of MCM8–9 loss, human cells lacking HROB showed a strong defect in DNA repair during DNA replication and HROB knockout mice were sterile, showing that, together with MCM8–9, HROB is important for genome maintenance and meiosis. We expect that elucidating HR, in which HROB and MCM8–9 are involved, would help cancer and fertility treatments in the future.
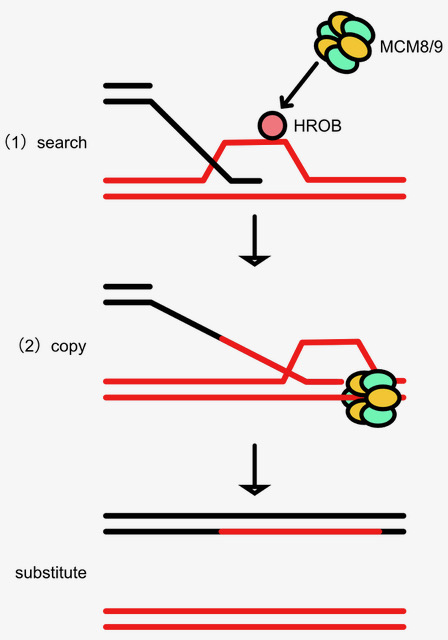
Figure: HR searches the donor sequence and uses DNA synthesis to copy and paste it to the recipient ones. In this paper, we found a novel factor, HROB, which promotes MCM8–9 loading to promote HR-associated DNA synthesis.
Message From NIGINTERN 2019
Neural signatures of sleep in zebrafish
Neural signatures of sleep in zebrafish.
Louis C Leung, Gordon X Wang, Romain Madelaine, Gemini Skariah, Koichi Kawakami, Karl Deisseroth, Alexander E Urban, and Philippe Mourrain
Nature 571(7764) 198-204 (2019) DOI:10.1038/s41586-019-1336-7
Slow-wave sleep and rapid eye movement (or paradoxical) sleep have been found in mammals, birds and lizards, but it is unclear whether these neuronal signatures are found in non-amniotic vertebrates. Here we develop non-invasive fluorescence-based polysomnography for zebrafish, and show—using unbiased, brain-wide activity recording coupled with assessment of eye movement, muscle dynamics and heart rate—that there are at least two major sleep signatures in zebrafish. These signatures, which we term slow bursting sleep and propagating wave sleep, share commonalities with those of slow-wave sleep and paradoxical or rapid eye movement sleep, respectively. Further, we find that melanin- concentrating hormone signalling (which is involved in mammalian sleep) also regulates propagating wave sleep signatures and the overall amount of sleep in zebrafish, probably via activation of ependymal cells. These observations suggest that common neural signatures of sleep may have emerged in the vertebrate brain over 450 million years ago.
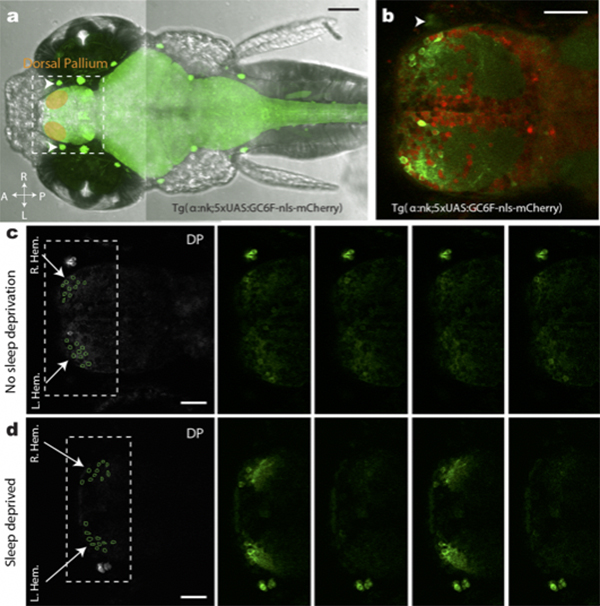
Figure: (a,b)Transgenic fish used for calcium imaging (c) Sleep non-deprived fish (d) slow bursting sleep seen in sleep deprived fish
▶This study is based on the previous study.
Glia-neuron interactions underlie state transitions to generalized seizures
Press release
Glia-neuron interactions underlie state transitions to generalized seizures
Carmen Diaz-Verdugo, Sverre Myren-Svelstad, Ecem Aydin, Evelien van Hoeymissen, Celine Deneubourg, Silke Vanderhaeghe, Julie Vancraeynest, Robbrecht Pelgrims, Mehmet LLyas Cosacak, Akira Muto, Caghan Kizil, Koichi Kawakami6, Nathalie Jurisch-Yaksi & Emre Yaksi
Nature Communications 10, Article number: 3830 (2019) DOI:10.1038/s41467-019-11739-z
Press release (In Japanese only)
Brain activity and connectivity alters drastically during the generalization of epileptic seizures. Throughout this transition, brain networks shift from a balanced resting state to a hyperactive and hypersynchronous state, spreading across the brain. It is however less clear which mechanisms underlie these state transitions. By studying neuronal and glia activity across the zebrafish brain, we observed striking differences between these networks. During pre-ictal period, neurons displayed a small increase in synchronous activity only locally, while the entire glial network was highly active and strongly synchronized across large distances. We observed that the transition from a pre-ictal state to a generalized seizure leads to an abrupt increase in neural activity and connectivity, which is accompanied by a strong functional coupling between glial and neuronal networks. Optogenetic activation of glia induced strong and transient burst of neuronal activity, emphasizing a potential role for glia-neuron connections in the generalization of epileptic seizures.
Sourse: Nature Communications 10, Article number: 3830 (2019)
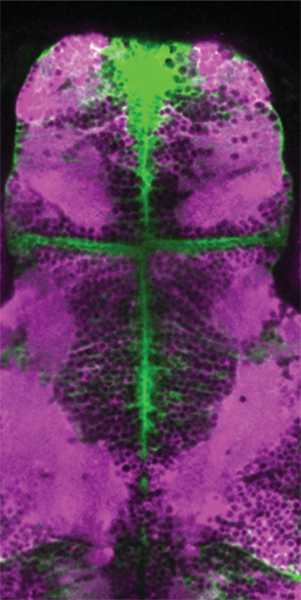
Fig: An image of the brain of the GFAP:Gal4;UAS:GCaMP6s;jRCaMP1a fish.
▶This study is based on the previous study.
The making of ‘Fancy Mouse’: Study reveals true cause of colorful hair on popular East Asian pet mice
Press release
Nested retrotransposition in the East Asian mouse genome causes the classical nonagouti mutation
Akira Tanave, Yuji Imai, Tsuyoshi Koide
Communications Biology 2 August 2019 10.1038/s42003-019-0539-7
EurekAlert! link about this artcle
For the past few hundred years, the colorful hair and unique patterns of the so-called “Fancy Mouse” have made them the stars of pet shows in Japan and beyond. Now, scientists have finally revealed the true cause of the genetic mutation responsible for the iconic black pigmentation in the popular East Asian pet.
Their findings were published on August 2, 2019 in Communications Biology. read more>

Fig: The mutated colors in Japanese fancy mice, JF1, gave them a Giant Panda look. The C57BL/6 (B6) strain has black coat color, nonagouti. Japanese wild strain MSM has agouti coat color.
*Credit: Tsuyushi Koide, the National Institute of Genetics (NIG) in Japan
▶ You can read the deeper story behind the paper below.
Reexamining the a of Mouse Genetics A to Z
▶Associate Prof. Koide posted “Comment” about this article.
A role for the rare endogenous retrovirus β4 in development of
Japanese fancy mice(PDF)
DFAST: Prokaryotic genome annotation pipeline for data submission to DDBJ
DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication
Yasuhiro Tanizawa, Takatomo Fujisawa, Yasukazu Nakamura Bioinformatics 34(6) 1037-9, 2018 DOI:10.1093/bioinformatics/btx713Current technology of high throughput sequencing forces microbiologists to deal with more and more genomic data than before. In addition, annotated genome data must be deposited in the public sequence database; however, its complicated process is an another burden for researchers. We have developed an automatic prokaryotic genome annotation pipeline called DDBJ Fast Annotation and Submission Tool (DFAST) aiming to assist data submission to DDBJ. DFAST is available as a web tool, which allows users to create data submission files using graphical user interface, and also as a stand-alone tool, which is well suited for massive data processing on a local machine. DFAST can annotate a typical-sized bacterial genome within 5 minutes and has unique features such as pseudogene detection. It is available at https://dfast.ddbj.nig.ac.jp.
DFAST will facilitate the use of genomic data in microbial studies.
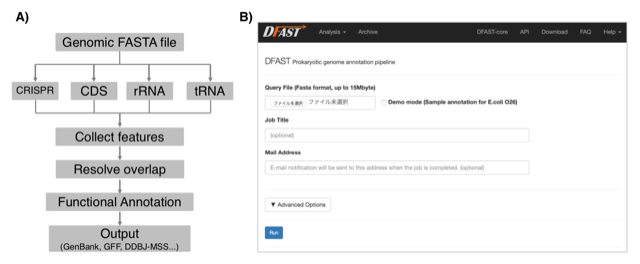
Figure: A) Annotation workflow of DFAST. B) Job submission form of the DFAST web server.
Visualization of autophagy in rice anther tapetum
Monitoring autophagy in rice tapetal cells during pollen maturation.
Shigeru Hanamata, Jumpei Sawada, Bunki Toh, Seijiro Ono, Kazunori Ogawa, Togo Fukunaga, Ken-Ichi Nonomura, Takamitsu Kurusu, Kazuyuki Kuchitsu.
Plant Biotechnology 36 (2): 99-105 (2019) DOI:10.5511/plantbiotechnology.19.0417a
Autophagy, a major catabolic pathway in eukaryotic cells, plays critical roles in the recycling of proteins and lipids, and is involved in many developmental processes. It plays important roles in pollen maturation and required for tapetal programmed cell death (PCD), postmeiotic anther development and nutrient supply to the pollens in rice (Kurusu et al. 2014). In this paper, we introduce an in vivo imaging technique to analyze the dynamics of autophagy in rice tapetum by expressing GFP-ATG8, a marker for autophagosomes under the control of tapetum-specific promoters. The results from imaging analyses including 3-dimensional visualization revealed that autophagy is dramatically induced at the specific stages throughout the tapetal cells just before their PCD process (Fig. 1). This monitoring system offers a powerful tool to analyze the regulation of autophagy in rice tapetum during anther development, and contribute to the research to increase the grain quality and yield of rice.
This work was supported in part by the NIG-Joint program (84A2018).
Glossary
Autophagosome: A spherical structure with double layer membranes. It is the key structure in macroautophagy, the intracellular degradation system for cytoplasmic contents.
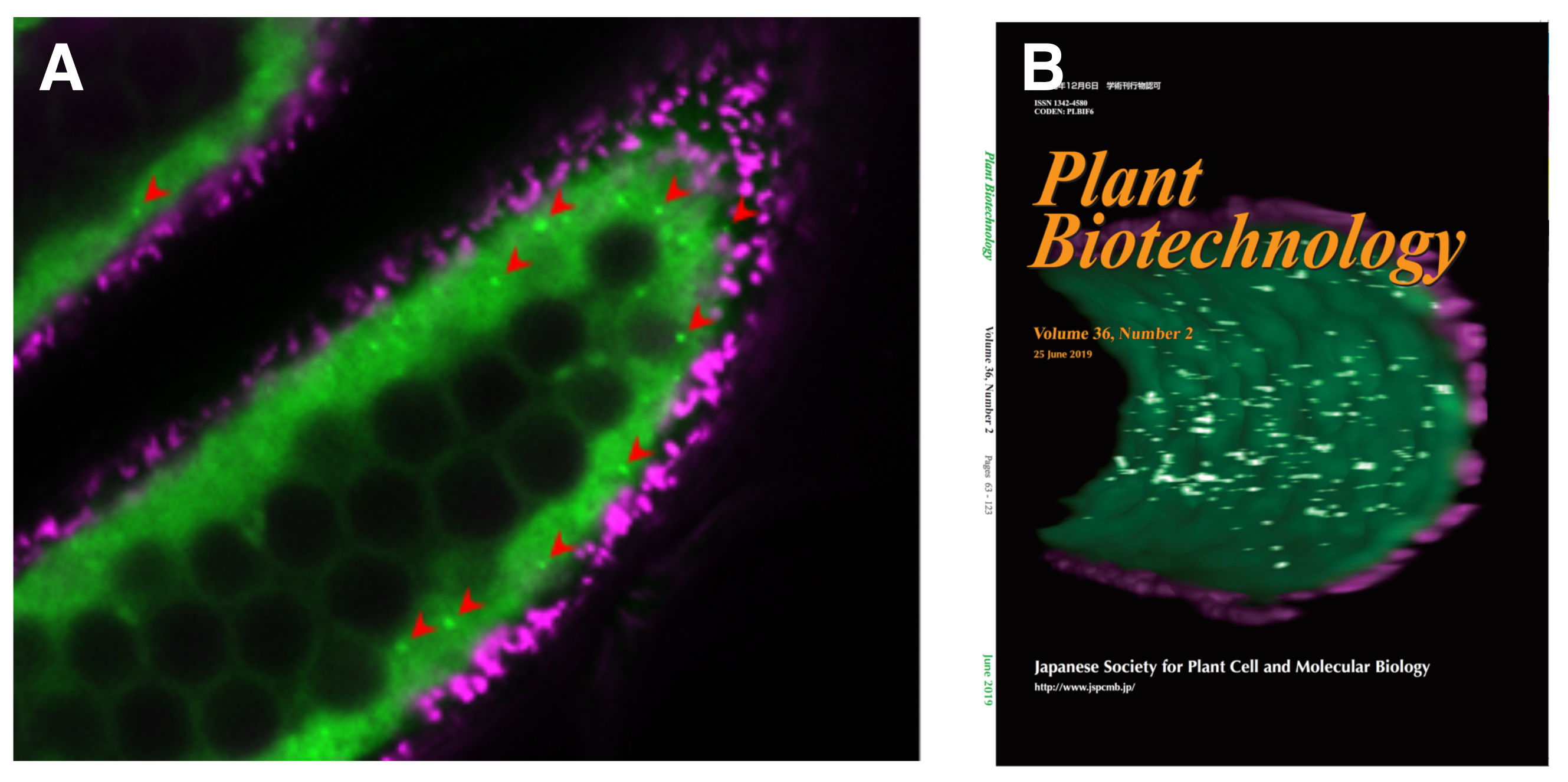
Figure: Visualization of the dynamics of autophagosomes/autophagy-related structures in rice anthers.
(A) A confocal fluorescence image of cross-section of rice anthers indicates GFP (green) and autofluorescence (magenta) of chlorophyll, respectively. Arrowheads indicate punctate signals of GFP-ATG8 (autophagosomes/autophagy-related structures). (B) Three-dimensional reconstruction of the tapetum undergoing autophagy.















