Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
NIG-GS (NIG Global Scholar) selection
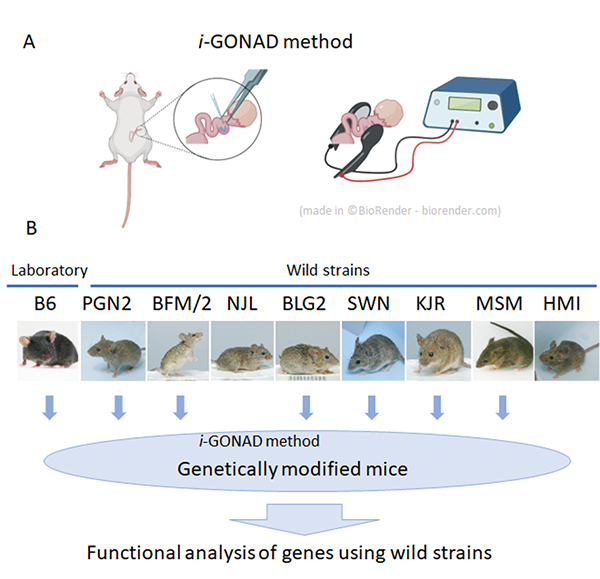
Genome editing is now possible in wild mouse strains!
Koide Group / Mouse Genomics Resource Laboratory
Efficient genome editing in wild strains of mice using the i-GONAD method
Yuji Imai, Akira Tanave, Makoto Matsuyama, and Tsuyoshi Koide
Scientific Reports (2022) 12, 13821 DOI:10.1038/s41598-022-17776-x
At the National Institute of Genetics, nine wild mouse strains have been established. These strains have characteristics not found in laboratory strains, such as marked genetic differences between different strains and behavior characteristic of wild mice.
These series of wild strains, named the Mishima Battery, are provided to researchers in Japan and overseas as highly unique resources, and are used for various research in the fields of cancer, immunology, development, and behavior. However, in spite of these excellent characteristics, wild strains have the problem of being difficult to apply genetic engineering technology.
In collaboration with Shigei Medical Research Institute and RIKEN, technical staff Yuji Imai and Associate Professor Tsuyoshi Koide of the Mouse Genomics Resource Laboratory have applied genome-editing using i-GONAD method, which does not involve any ex vivo manipulation of unfertilized or fertilized egg. The group showed that it is possible to efficiently modify genes in most wild strains by using this method.
First, in vitro fertilization was performed on the experimental strain C57BL/6 (B6 strain) and nine wild strains to investigate the efficiency of in vitro fertilization necessary for general genome editing experiments. As a result, it was found that only two wild strains were able to perform in vitro fertilization with the similar efficiencies as the B6 strain, and the other strains were extremely inefficient. Therefore, we applied a method called i-GONAD, developed by Professor Masato Otsuka of Tokai University, to wild strains, which performs genome editing without ex vivo manipulation of fertilized eggs. As a result, we succeeded in performing genome editing in 7 out of the 9 strains examined. This result indicates that it has become possible to efficiently perform genetic modification using wild strains in the future, and the use of wild strains in many research fields can be expected.
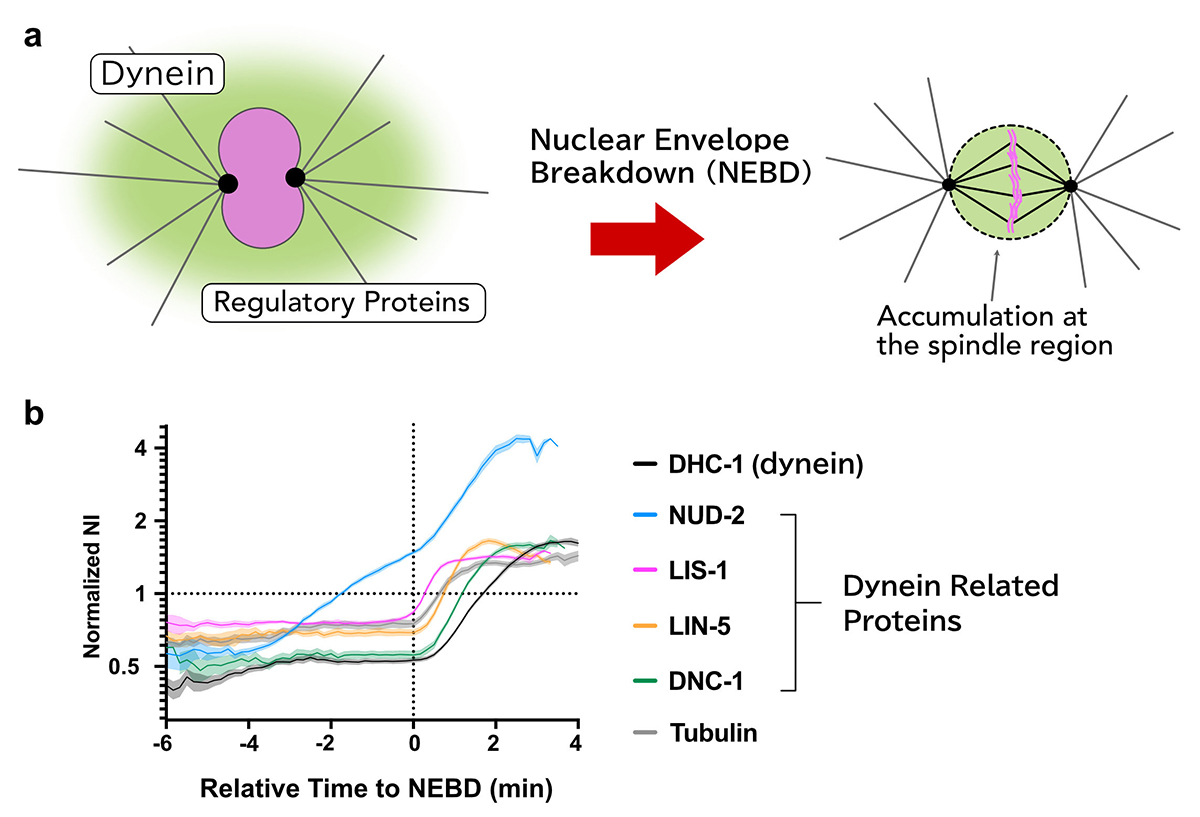
Sequential accumulation of dynein and its related proteins during mitosis and a possible sequential activation mechanism
Kimura Group / Cell Architecture Laboratory
Sequential accumulation of dynein and its regulatory proteins at the spindle region in the Caenorhabditis elegans embryo
Takayuki Torisawa, & Akatsuki Kimura
Scientific Reports (2022) 12, 11740 DOI:10.1038/s41598-022-15042-8
Cytoplasmic dynein is a molecular motor responsible for various cellular activities, including intracellular transport and cell division. To achieve various functions, dynein needs to be regulated by other proteins, and it has still been elusive how these regulations are achieved in many cellular contexts.
Using the early embryos of Caenorhabditis elegans, we focused on the spatiotemporal regulation of dynein during mitosis, where dynein and its regulatory proteins translocated from the cytoplasm to the spindle region, and observed the dynamics of dynein and the regulatory proteins.
We revealed that there are i) selectivity, ii) varieties in the accumulation sites, and iii) the order of accumulation in the accumulation dynamics in the spindle region.
Furthermore, we found that the accumulation of NUD-2 was unique among the dynein regulators we analyzed. NUD-2 started to accumulate before NEBD (pre-NEBD accumulation). Using a protein injection approach, we revealed that the C-terminal helix of NUD-2 was responsible for pre-NEBD accumulation. These findings suggest a fine temporal control of the subcellular localization of regulatory proteins.
This work was supported by JSPS Grants-in-Aid for Scientific Research JP19K16094, JP18H02414, JP18H05529, and JP18KK0202.
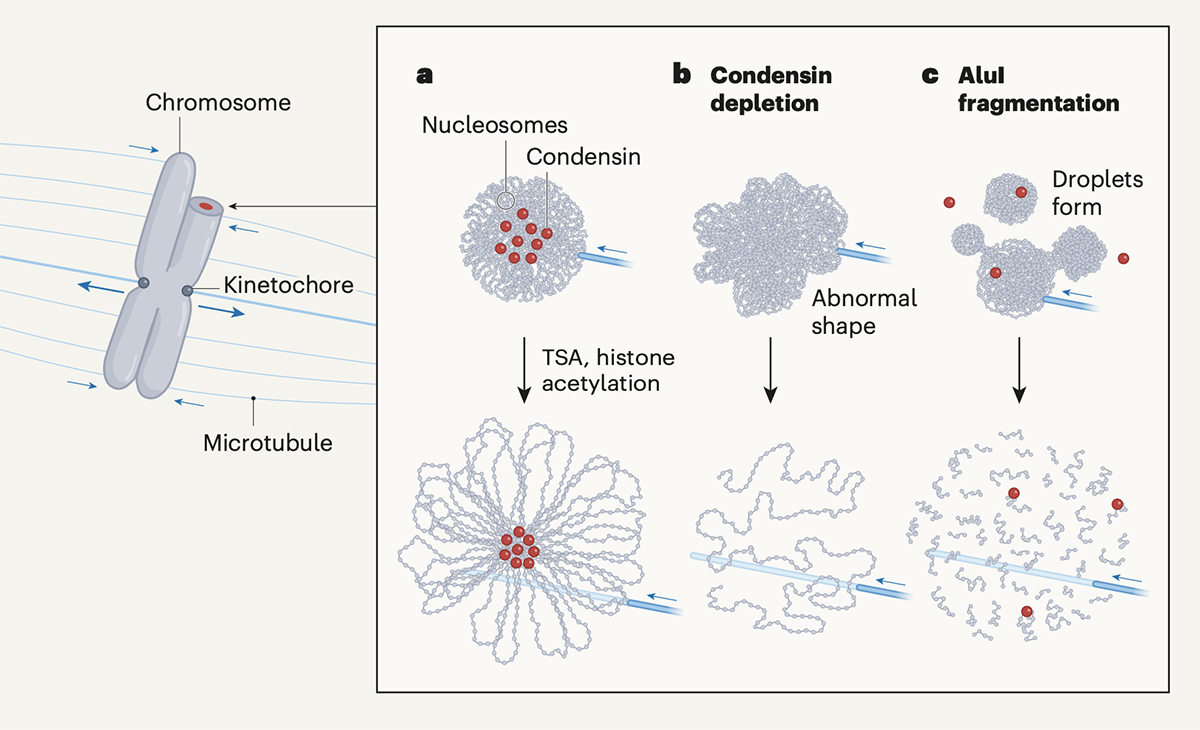
A phase transition for chromosome transmission when cells divide
Maeshima Group / Genome Dynamics Laboratory
A phase transition for chromosome transmission when cells divide
Kazuhiro Maeshima
Nature 2022 August 03 DOI:10.1038/d41586-022-01925-3
Mitotic chromosomes are the structure where DNA is tightly compacted, and they carry genetic information to be passed on to the next generation. These chromosomes are transmitted from the mother cell onto the daughter cell with physical force from microtubes and other factors. In other words, chromosomes should have mechanical resistance to endure such forces.
Recently, Daniel W. Gerlich and his colleagues have shown that chromosomes condense and gain such mechanical resistance by phase transition(Schneider et al. “A chromatin phase transition protects mitotic chromosomes against microtubule perforation” Nature 2022 doi: 10.1038/ s41586-022-05027-y). In this paper, Schneider et al. have shown that global histone deacetylation cause phase transition in mitotic chromosomes, which makes them condensed and resistant to mechanical forces. The authors have also shown that a protein complex called condensin, which was previously thought to be essential for chromosome condensation, is not involved in the condensation process itself.
Professor Kazuhiro Maeshima at Genome Dynamics Laboratory wrote a commentary on this paper in the News & Views section of Nature. Maeshima discussed how global histone deacetylation causes chromosome condensation and the role of condensin in shaping rod-like chromosomes (chromatin loop formation mechanism).
Change of publication location of new coronavirus infections at our institute
July 25, 2022
Research Organization of Information and Systems
National Institute of Genetics
The announcement of new coronavirus infections after July 25 will be made on the following website instead of the Information section.
NIG Measures for the Novel Coronavirus Infectious Disease
Summer Holiday (Aug. 15-16)
NIG will be closed from August 15 to August 16, 2022 for summer holiday.
Thank you for your understanding and cooperation.
Faculty member MURAYAMA at the Center for Frontier Research has been awarded tenure
MURAYAMA, Yasuto tenure track associate professor at the Center for Frontier Research, has been awarded tenure as of July 1, 2022.
MURAYAMA, Yasuto: Chromosome Biochemistry Laboratory

- MURAYAMA, Yasuto
Associate Professor
New assistant professor joins NIG
New assistant professor joins NIG as of July 1, 2022.
KAWAGUCHI,Akane : Kuraku Group • Molecular Life History Laboratory
Three genes instrumental in increasing harvests in the earlier step of rice domestication
A stepwise route to domesticate rice by controlling seed shattering and panicle shape
Ryo Ishikawa, Cristina Cobo Castillo, Than Myint Htun, Koji Numaguchi, Kazuya Inoue, Yumi Oka, Miki Ogasawara, Shohei Sugiyama, Natsumi Takama, Chhourn Orn, Chizuru Inoue, Ken-Ichi Nonomura, Robin Allaby, Dorian Q Fuller and Takashige Ishii
PNAS (2022) 119, e2121692119 DOI:10.1073/pnas.2121692119
![]() Press release (In Japanese only)
Press release (In Japanese only)
The international collaborative group of Kobe University, National Institute of Genetics (NIG), University of Warwick, University College London, Yezin Agricultural University, Cambodian Agricultural Research and Development Institute, successfully unveiled that the Asian wild rice (O. rufipogon Griff.) dramatically had increased grain yields with mutations at three genes in an initial step of domestication.
The loss of seed-shattering behaviour is an important step of domestication enabling humans to harvest more grains. At first, to experimentally reproduce an earlier process of rice domestication, several domestication-related genes were replaced with those of cultivated rice in the genetic background of wild rice, O. rufipogon. The replacement alone with a cultivated rice-type mutation of sh4, a rice gene with a major impact on seed shattering, was insufficient, but the replacement with cultivated rice-type mutations of both sh4 and qSH3, an allele within the seed-shattering gene OsSh1, caused the partial loss of abscission layer formation in O. rufipogon. However, the combination of two mutations of sh4 and qSH3 was not sufficient to increase yield under natural conditions. To further increase yield, our group found that the replacement together with cultivated rice-type mutations of three genes, sh4, qSH3 and SPR3, dramatically increased yields (Fig. 1). SPR3 is a rice gene controlling a panicle shape, and the closed panicle property enhances long awn-retaining seeds entangled, and in addition, reduces a bending moment which is a predominant factor affecting seed dispersal (Fig. 2), resulting in increased harvests.
We propose a stepwise route in the earliest phase of rice domestication, in which SPR3-controlled closed panicle morphology was instrumental in addition to sequential recruitments of sh4- and qSH3-dependent loss of shattering.
This study was supported partly by NIG-JOINT program 83A2016-2018. The wild rice accession, maintained by NIG in the support of National Bioresource Project (NBRP) Rice, was used in this study.
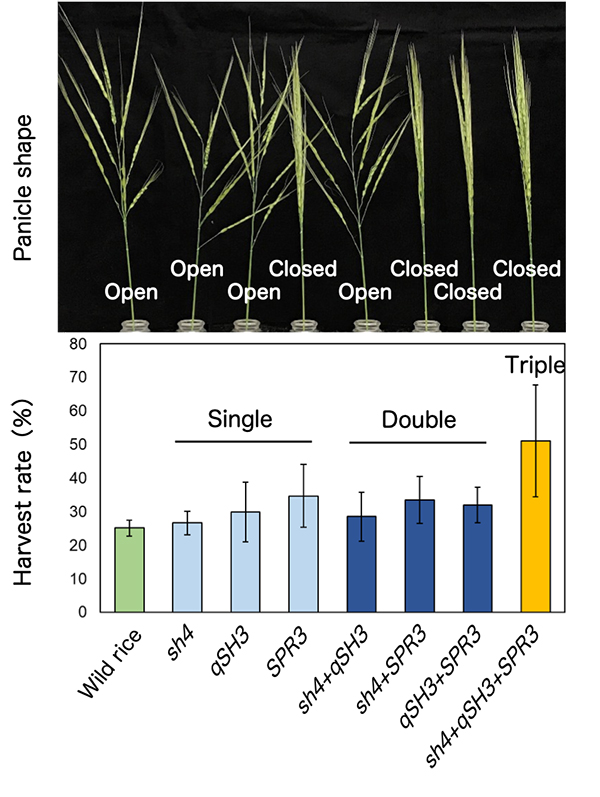
(Top) The panicle shape of seven wild rice introgression lines with different replacements to cultivated rice-type alleles at sh4, qSH3 and SPR3. (bottom) The seed harvest rate of respective wild rice introgression lines.
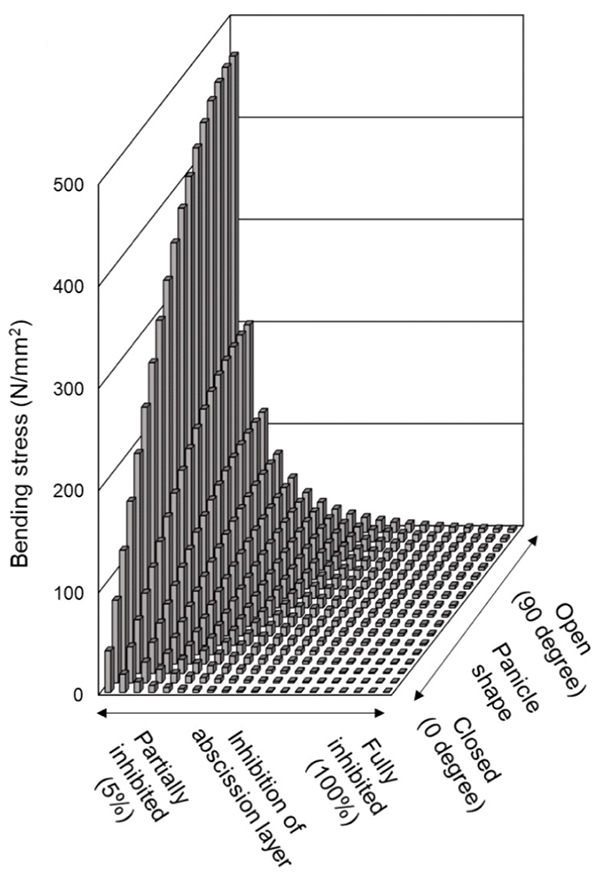
A higher stress was experienced in the open panicle with less inhibited abscission. This simulation suggests that in addiotion to the replacement with sh4 and qSH3 for reduced shuttering, we propose that the replacement with SPR3 for panicles closing might contribute to increased harvest rate during an earlier step of rice domestication.
Detection of Ancient Viruses and Long-Term Viral Evolution
Inoue Group / Human Genetics Laboratory
Detection of Ancient Viruses and Long-Term Viral Evolution
Luca Nishimura, Naoko Fujito, Ryota Sugimoto, and Ituro Inoue*
Viruses (2022) 14, 1336 DOI:10.3390/v14061336
Archaeological remains contain Ancient DNA and RNA, and those nucleic acids provide genomic information about ancient people together with ancient microbiomes and viruses that infected ancient individuals. Since 1997, more than 20 ancient viral species including influenza virus and hepatitis B virus have been discovered from historical samples such as bones and mummified tissues. Those ancient viral genomes have been utilized to estimate the past pandemics of pathogenic viruses within the ancient human population and long-term evolutionary events. In our review article, we overview the ancient viral studies and experimental and analytical techniques and discuss the long-term viral evolutionary studies using ancient viral genomes.
Chromatin behavior in living cells: lessons from single-nucleosome imaging and tracking
Maeshima Group / Genome Dynamics Laboratory
Chromatin behavior in living cells: lessons from single-nucleosome imaging and tracking
*Satoru Ide, Sachiko Tamura, *Kazuhiro Maeshima *corresponding author
BioEssays 2022 June 03 DOI:10.1002/bies.202200043
Eukaryotic genome DNA is wrapped around core histones and forms a nucleosome structure. Together with associated proteins and RNAs, these nucleosomes are organized three-dimensionally in the cell as chromatin. Emerging evidence demonstrates that chromatin consists of rather irregular and variable nucleosome arrangements without the regular fiber structure and that its dynamic behavior plays a critical role in regulating various genome functions. Single-nucleosome imaging is a promising method to investigate chromatin behavior in living cells. It reveals local chromatin motion, which reflects chromatin organization not observed in chemically fixed cells. The motion data is like a gold mine. Data analyses from many aspects bring us more and more information that contributes to better understanding of genome functions. In this review article, we describe imaging of single-nucleosomes and their tracked behavior through oblique illumination microscopy. We also discuss applications of this technique, especially in elucidating nucleolar organization in living cells.
This work was supported by JSPS Grants and MEXT KAKENHI grants (21H02535, 20H05936 and 21H02453) and the Uehara Memorial Foundation.
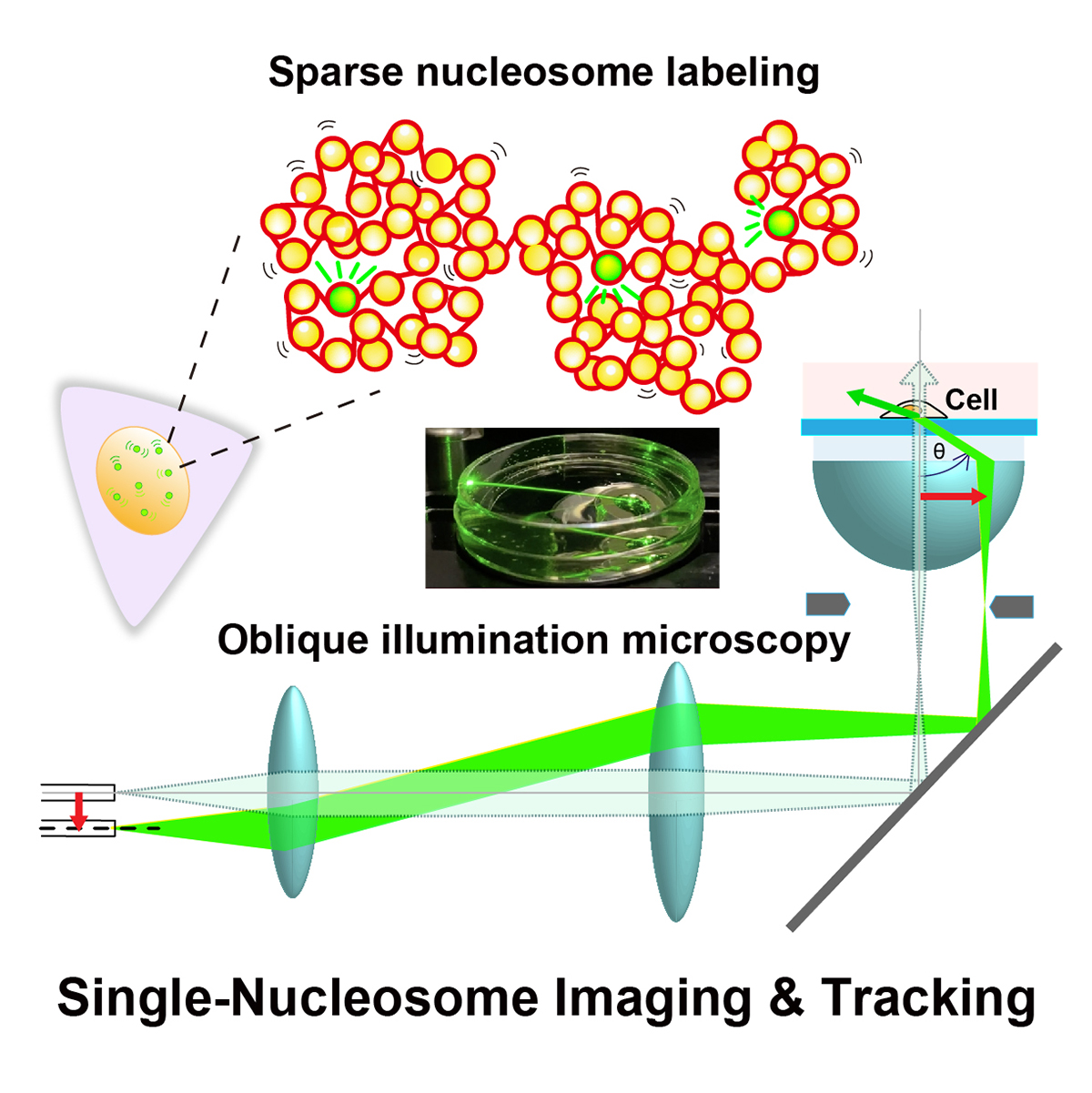
Figure: Single-nucleosome imaging and tracking is a promising technique to investigate dynamic chromatin behavior in living cells. We describe how this imaging works using sparse nucleosome labeling and oblique illumination microscopy, and how information on chromatin dynamics can be extracted from the obtained motion data.
Video1: How laser beams traveled through a glass dish filled with medium using an oblique illumination microscopy. As the incident laser beam shifts from the center axis, higher refraction angles of the beam from the glass are obtained. At the last 2 frames, total internal reflection (TIR) occurs and only the glass surface is illuminated.
Video2: Movie data (50 ms/frame) of single nucleosomes (basic units of chromatin) fluorescently labeled in a living human cell. Note that clear, well-separated dots and their movements were visualized.
Steady-state chromatin motion throughout interphase
Press release
Single-nucleosome imaging reveals steady-state motion of interphase chromatin in living human cells
Shiori Iida, Soya Shinkai, Yuji Itoh, Sachiko Tamura, Masato T. Kanemaki, Shuichi Onami, Kazuhiro Maeshima
Science Advances (2022) 8, eabn5626 DOI:10.1126/sciadv.abn5626
![]() Press release (In Japanese only)
Press release (In Japanese only)
The human body is composed of over forty trillion cells. Each of these cells has totally two meters of tightly packaged genomic DNA, the blueprint of life. Recently, there have been many advances in understanding how DNA is packaged and organized as chromatin in the cell. In contrast, how chromatin behaves in living cells remains unclear.
SOKENDAI graduate student Shiori Iida, JSPS Fellow Yuji Itoh, Technical Stuff Sachiko Tamura, and Professor Kazuhiro Maeshima of Genome Dynamics Laboratory (NIG), together with Research Scientist Soya Shinkai and Team Leader Shuichi Onami of RIKEN BDR, and Professor Masato T. Kanemaki of Molecular Cell Engineering Laboratory (NIG), have investigated the local movements of chromatin in living human cells using super-resolution fluorescence microscopy (Movie 1).
Both DNA amount and nuclear size become double during the preparation period for the cell division (interphase). Previously, it has been suggested that these drastic changes in the nuclear environment would affect chromatin movements. However, Iida et al. have revealed that chromatin motion keeps a steady state throughout the interphase. Chromatin motion is directly related to the accessibility of the DNA (readability of genomic information). Steady-state chromatin motion allows cells to conduct housekeeping tasks under similar environments during interphase.
This work was supported by JSPS and MEXT KAKENHI grants (20H05550、21H05763、19K23735、 20J00572、18H05412、19H05273、20H05936), a Japan Science and Technology Agency CREST grant (JPMJCR15G2), JST SPRING(JPMJSP2104), the Takeda Science Foundation, and the Uehara Memorial Foundation.
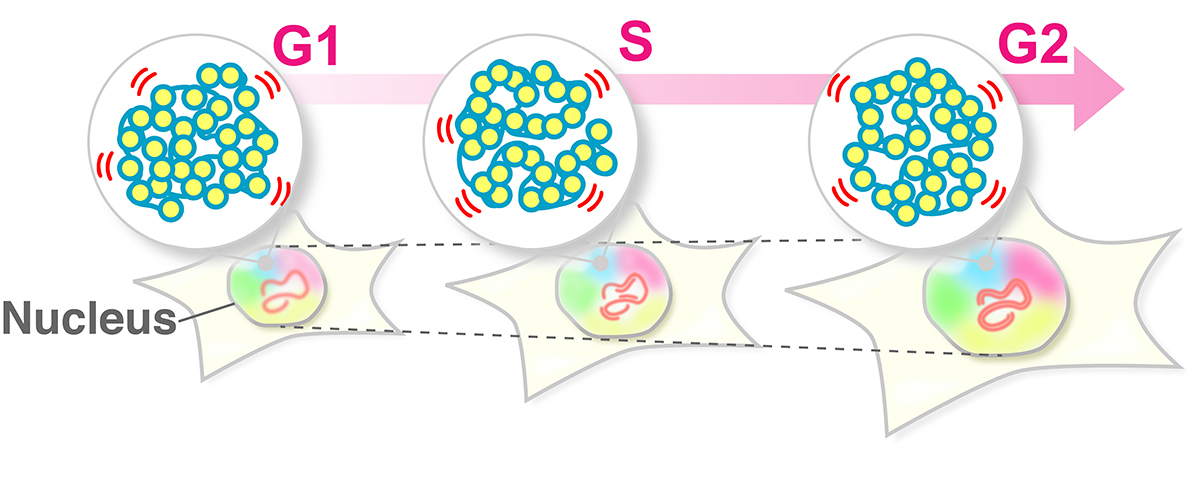
Figure: The swaying motion of chromatin keeps a steady state throughout the interphase (G1, S, G2 phases) despite increases in DNA amount and nuclear volume. Steady-state chromatin motion allows cells to conduct housekeeping tasks under similar environments during interphase.
Video1: Movie data (50 ms/frame) of single nucleosomes (basic units of chromatin) fluorescently labeled in a living human cell. Note that clear, well-separated dots and their movements were visualized.
Video2: Movie data (50 ms/frame) of single nucleosomes fluorescently labeled in living human cells; (left) G1-phase, (right) G2-phase. Note that there is not much difference in nucleosome motion between G1 and G2 phase, even as nuclear size increases.
▶ The article of EurekAlert! is here.
The egg is not a simple ellipsoid
Press release
The extra-embryonic space and the local contour are critical geometric constraints regulating cell arrangement
*Sungrim Seirin-Lee, Kazunori Yamamoto, *Akatsuki Kimura
*Corresponding authors
Development (2022) 149, dev200401 DOI:10.1242/dev.200401
![]() Press release (In Japanese only)
Press release (In Japanese only)
Arrangement of cells which defines how cells contact each other is important in developmental processes. The mechanisms determining cell arrangement can be classified into three factors: orientation of cell division, interaction between cells, and geometrical constraints provided by surrounding structures where cells are confined such as the eggshell. Among these factors, the contribution of geometrical constrains have been less explored. Many of theoretical and experimental approaches exploring cell arrangements inside the eggshell have assumed the eggshell as a pure ellipsoidal shape. In this study, the authors developed a computational model based on a phase-field method that can incorporate the real shape of the eggshells. Using this model and the experimental data obtained with the nematode Caenorhabditis elegans embryo, the authors showed that an arrangement observed in vivo can be explained by the real shape but not by an ellipsoid. Extending this finding, the authors found the amount of the extra-embryonic space (ES), the empty space within the eggshell not occupied by embryonic cells, is critical to define cell arrangement. The study proposed that the local features of geometric constraints such as the ES, play important roles in cell arrangement, which should be important for any multicellular systems.
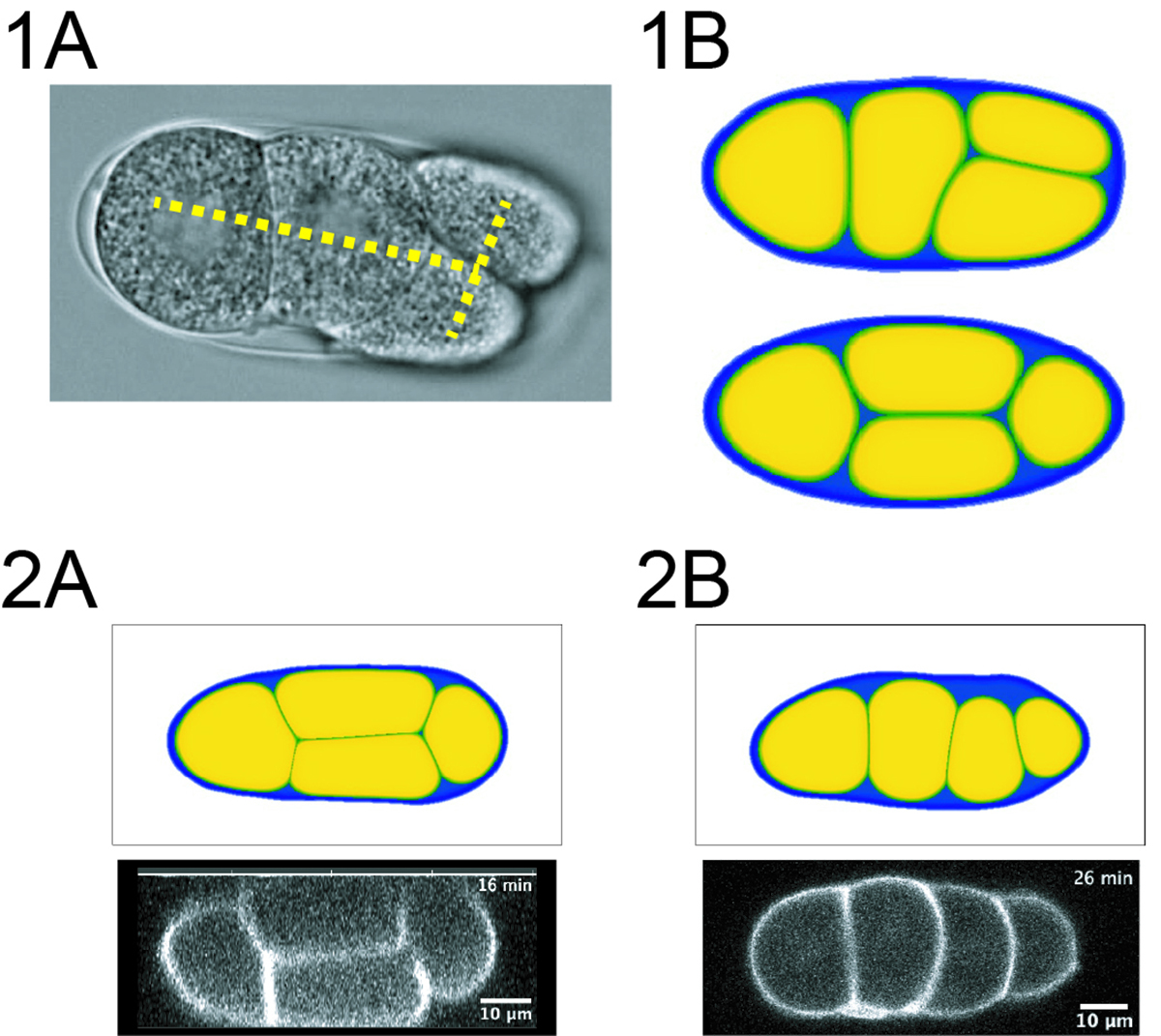
1B) A simulation reproducing the T-reverse arrangement. (Upper) A simulation incorporating the real shape of the eggshell reproduced the T-reverse arrangement. (Lower) A simulation with an ellipsoidal eggshell did not show the T-reverse arrangement, but a “Diamond” arrangement.
2) Increasing the amount of the ES changed the cell arrangement. 2A) In the case of ES=10%. The diamond arrangement was observed in simulation (upper) and in real embryo (lower).
2B) In the case of ES=20%. A linear arrangement was observed in simulation (upper) and in real embryo (lower).
Prof. Kanemaki’s News and Views was published in Nature. “A rethink about enzymes that drive DNA replication”
Prof. Kanemaki’s News and Views on the recent finding on DNA replication was published in Nature.
A rethink about enzymes that drive DNA replication
Comment from Prof. Kanemaki:
I wrote a short review article about a new paper published in Nature. It has been believed that two kinases, CDC7 and CDK2, drive DNA replication. However, this paper showed that either CDC7 or CDK1 suffices for driving DNA replication in mouse and human cells. In addition, the auxin-inducible degron (AID) that my group developed was used in this research.
Kanemaki Group • Molecular Cell Engineering Laboratory
Cell Nucleus issue of Current Opinion in Cell Biology edited by Prof. Maeshima has been published.

A special issue on Cell Nucleus of Current Opinion in Cell Biology edited by Prof. Kazuhiro Maeshima at Genome Dynamics Laboratory and Prof. Eran Meshorer, Hebrew University of Israel, has been published.
There are 11 review articles on cell nuclei in Vol. 74, published in February 2022, and 4 articles in Vol. 75, published in April 2022.
https://www.sciencedirect.com/journal/current-opinion-in-cell-biology/vol/74/suppl/C
https://www.sciencedirect.com/journal/current-opinion-in-cell-biology/vol/75/suppl/C
Vol. 74 includes “Ligand-induced degrons for studying nuclear functions” by NIG Prof. Masato Kanemaki and also a review paper on plant chromatin organization by NIG Visiting Professor Frederic Berger.
https://www.sciencedirect.com/science/article/pii/S0955067421001228
https://www.sciencedirect.com/science/article/pii/S0955067421001186
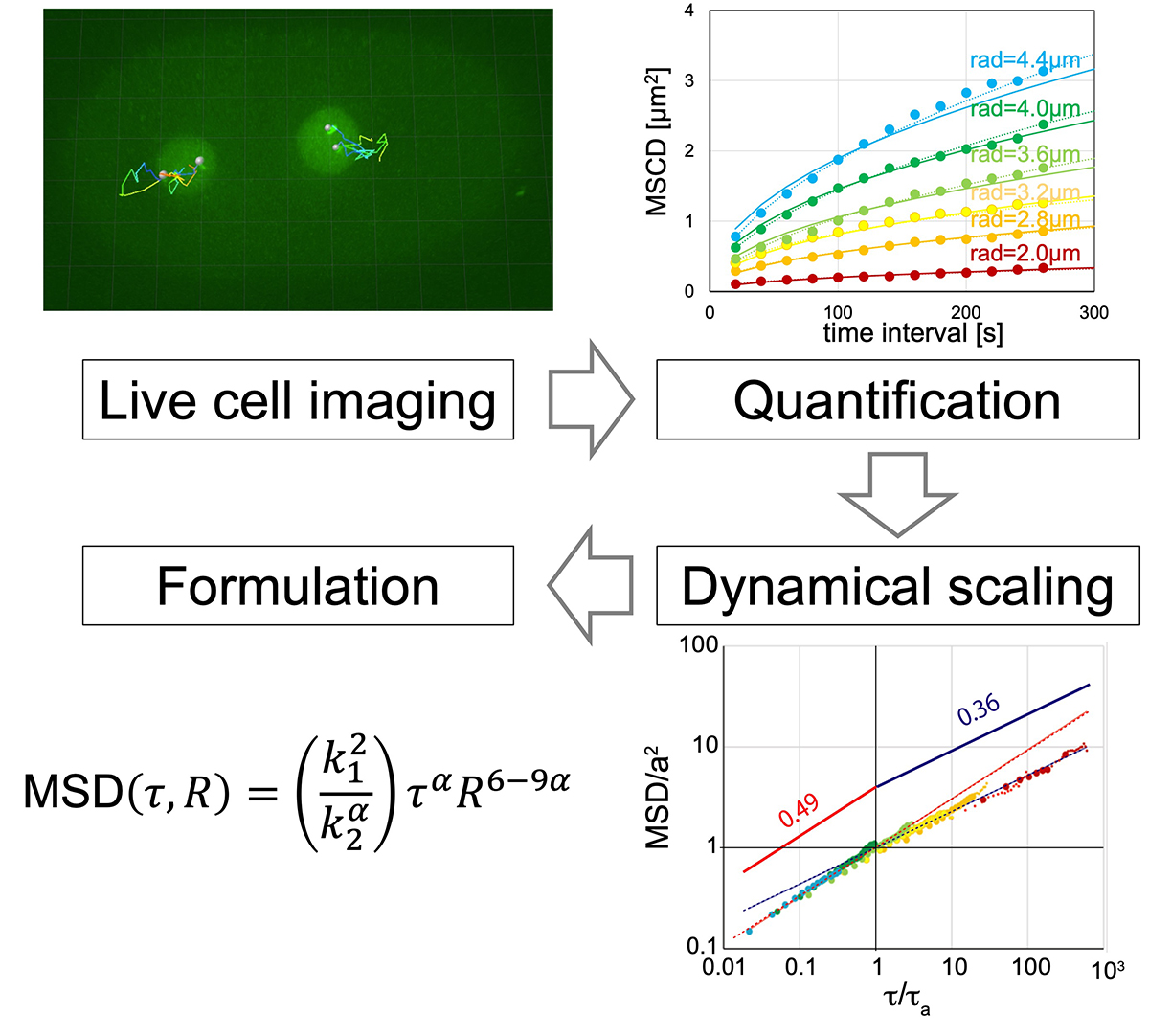
How do genes move in living cells?
Press release
Formulation of chromatin mobility as a function of nuclear size during C. elegans embryogenesis using polymer physics theories.
Aiya K. Yesbolatova, Ritsuko Arai, *Takahiro Sakaue, and *Akatsuki Kimura. *Corresponding authors
Physical Review Letters (2022) 128, 178101 DOI:10.1103/PhysRevLett.128.178101
![]() Press release (In Japanese only)
Press release (In Japanese only)
By combining theory and experiment, Aiya K. Yesbolatova and her collaborators provided unambiguous evidence that the chromatin in early embryos obeys the universal dynamics predicted by the polymer physics. Although the chromatin dynamics is believed to play a key role in controlling the gene expression, its quantitative characterization has been elusive mainly due to the complexity in living cells. The findings and formulation help researchers quantify the chromatin motion in living cells, thus laying the foundation for future research in this biologically important problem.