Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
A mechanism of cell fate maintenance through histone acetylation
Multicellular Organization Laboratory • Sawa Group
HTZ-1/H2A.z and MYS-1/MYST HAT act redundantly to maintain cell fates in somatic gonadal cells through repression of ceh-22 in C. elegans
Yukimasa Shibata, Hitoshi Sawa and Kiyoji Nishiwaki
Development 141, 209-218 (2013), doi:10.1242/dev.090746
The stable maintenance of cell fates is essential for proper development and tissue homeostasis. Abnormalities in the maintenance lead to cancer or loss of important cell types. We have previously shown histone acetyltransferases (MYS-1 and MYS-2) and an acetylated-histone binding protein (BET-1) are required for the maintenance of cell fates in C. elegans, indicating that histone acetylation plays important roles in the process. It is not known, however, how BET-1 maintains cell fates.
It is known, in yeast, that BDF1 (an homolog of BET-1) forms a complex with a chromatin-remodeling factor SWR1 and regulates the deposition of histone H2A variant HTZ1/H2A.z. We have shown that SSL-1/SWR1 and HTZ-1/H2A.z are required for the cell fate maintenance in C. elegans. These proteins suppress the ectopic production of germline niche cells (DTC) by maintaining fates of somatic cells in the gonad. We found that the ceh-22 gene that encodes a transcription factor required for the DTC production is a direct target of HTZ-1 and that ceh-22 transcription is repressed by BET-1, MYS-1 and HTZ-1. Although it is well known that histone acetylation is involved in transcriptional activation, our results indicate that it also represses transcription.
This study has been carried out as collaboration with Drs. Shibata and Nishiwaki at Kwansei Gakuin University.
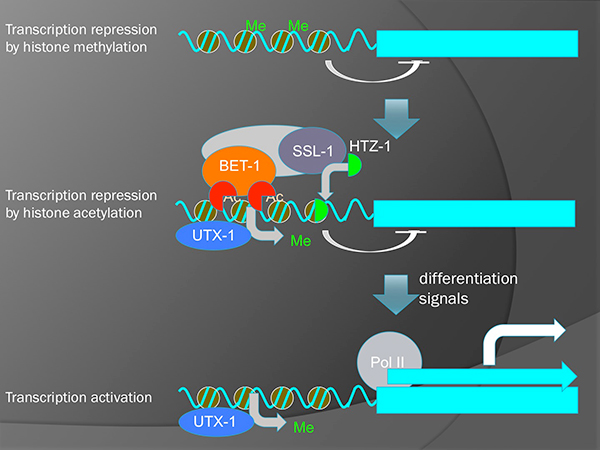
A model for two-step activation of transcription. Removal of H3K27 methylation by UTX-1 potentiates genes to be transcribed. However, actual transcription is still inhibited by BET-1 and HTZ-1 without the reception of differentiation signals.
Centoromere specific chromatin structure
Division of Molecular Genetics • Fukagawa Group
The centromeric nucleosome-like CENP–T–W–S–X complex induces positive supercoils into DNA
Kozo Takeuchi, Tatsuya Nishino, Kouta Mayanagi, Naoki Horikoshi,, Akihisa Osakabe, Hiroaki Tachiwana, Tetsuya Hori, Hitoshi Kurumizaka and Tatsuo Fukagawa
Nucleic Acids Research, (2013), doi:10.1093/nar/gkt1124
The centromere is a specific genomic region upon which the kinetochore is formed to attach to spindle microtubules for faithful chromosome segregation. To distinguish this chromosomal region from other genomic loci, the centromere contains a specific chromatin structure including specialized nucleosomes containing the histone H3 variant CENP-A. In addition to CENP-A nucleosomes, we have found that centromeres contain a nucleosome-like structure comprised of the histone-fold CENP-T-W-S-X complex (Nishino et al., Cell, 2012). However, it is unclear how the CENP-T-W-S-X complex associates with centromere chromatin. Here, we demonstrate that the CENP-T-W-S-X complex binds preferentially to ~100 bp of linker DNA rather than nucleosome-bound DNA. In addition, we find that the CENP-T-W-S-X complex primarily binds to DNA as a (CENP-T-W-S-X)2 structure. Interestingly, in contrast to canonical nucleosomes that negatively supercoil DNA, the CENP-T-W-S-X complex induces positive DNA supercoils. We found that the DNA-binding regions in CENP-T or CENP-W, but not CENP-S or CENP-X, are required for this positive supercoiling activity and the kinetochore targeting of the CENP-T-W-S-X complex. In summary, our work reveals the structural features and properties of the CENP-T-W-S-X complex for its localization to centromeres.
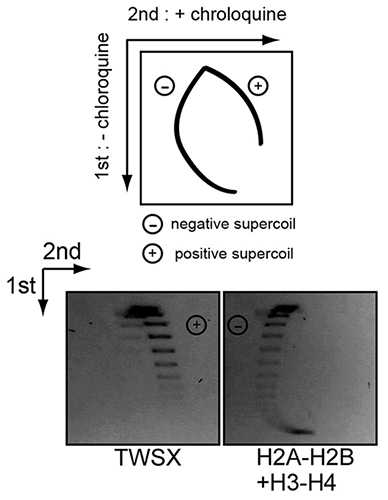
Experimental design to distinguish negative and positive supercoils (top). Negative supercoiled topoisomers slowly migrate following addition of chloroquine, whereas positive supercoiled topoisomers migrate faster following chloroquine addition. Plasmid supercoiling assays performed with CENP-T-W-S-X and histone H2A-H2B-H3-H4 complexes in the presence or absence of chloroquine (Below). The CENP-T-W-S-X induces positive supercoils.
Development of a New Method for Synthesis of Tandem Hairpin Pyrrole−Imidazole Polyamide Probes Targeting Human Telomeres
Biological Macromolecules Laboratory (Maeshima Group)
Development of a New Method for Synthesis of Tandem Hairpin Pyrrole–Imidazole Polyamide Probes Targeting Human Telomeres
Kawamoto, Y., Bando, T. *, Kamada, F., Li, Y., Hashiya, K., Maeshima, K. *, and Sugiyama, H. *
*co-corresponding authors
Journal of the American Chemical Society (JACS) October 1, 2013 DOI:10.1021/ja406737n
Pyrrole–imidazole (PI) polyamides bind to the minor groove of DNA in a sequence-specific manner without causing denaturation of DNA. To visualize telomeres specifically, tandem hairpin PI polyamides conjugated with a fluorescent dye have been synthesized, but the study of telomeres using these PI polyamides has not been reported because of difficulties synthesizing these tandem hairpin PI polyamides. To synthesize tandem hairpin polyamides more easily, we have developed new PI polyamide fragments and have used them as units in Fmoc solid-phase peptide synthesis. Using this new method, we synthesized four fluorescent polyamide probes for the human telomeric repeat TTAGGG. The polyamides synthesized using the new method successfully targeted to human and mouse telomeres under a physiological condition and allow easier labeling of telomeres in the cells while maintaining the telomere structure. Using the fluorescent polyamides, we demonstrated that the telomere length at a single telomere level is related to the abundance of TRF1 protein, a shelterin complex component in the telomere.
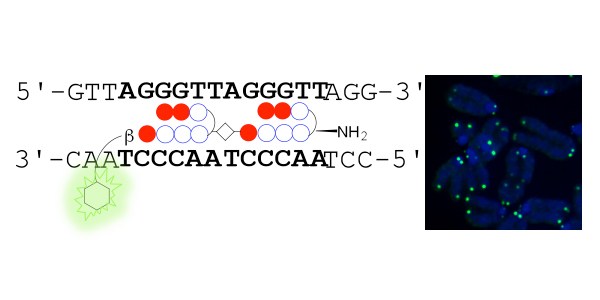
Left, schematic representation showing that TH59 compound with a fluorescent dye binds to the human telomere sequence (TTAGGG repeat). Right, the telomere regions of chromosome ends are labeled with TH59 (green). DNA stain (blue).
Chromatin compaction protects genomic DNA from radiation damage
Press release
Chromatin compaction protects genomic DNA from radiation damage
Takata, H., Hanafusa, T., Mori T., Shimura, M., Iida, Y., Ishikawa, K., Yoshikawa, K., Yoshikawa, Y., Maeshima, K.
PLOS ONE 8(10): e75622. doi:10.1371/journal.pone.0075622
Genomic DNA is three-dimensionally organized into the nucleus, and is thought to form compact chromatin domains. Although chromatin compaction is known to be essential for mitosis, whether it confers other advantages, particularly in interphase cells, remains unknown. Here, we report that chromatin compaction protects genomic DNA from radiation damage. Using a newly developed solid-phase system, we found that the frequency of double-strand breaks (DSBs) in compact chromatin after ionizing irradiation was 5-50-fold lower than in decondensed chromatin. Since radical scavengers inhibited DSB induction in the decondensed chromatin, the condensed chromatin had a lower level of reactive radical generation after ionizing irradiation. We show that chromatin compaction also protects DNA from attack by chemical agents. Our findings suggest that genomic DNA compaction plays an important role in maintaining genomic integrity.
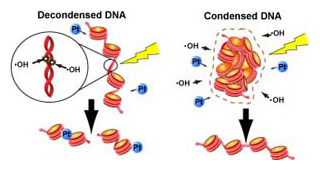
DNA condensation with fewer water molecules (right: condensed DNA) that there is less risk of being attacked by hydroxyl radicals. The situation is also effective to protect DNA from the binding of cisplatin (Pt).
Regulation of glycinergic synapse and behavior by a RNA helicase
Motor Neural Circuit Laboratory • Hirata Group
Hirata, H., Ogino, K., Yamada, K., Leacock, S. and Harvey, R. J.
J. Neurosci. 33: 14638-14644 (2013). doi: 10.1523/JNEUROSCI.1157-13.2013
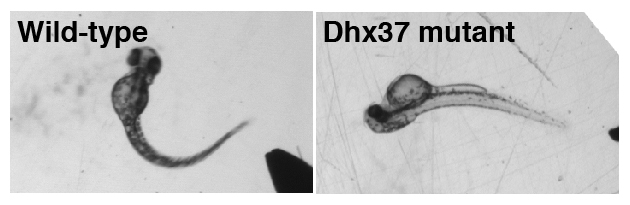
RNA helicases regulate RNA metabolism, but their substrate specificity and in vivo function remain largely unknown. We isolated spontaneous mutant zebrafish that exhibit an abnormal dorsal bend at the beginning of tactile-evoked escape swimming. Similar behavioral defects were observed in zebrafish embryos treated with strychnine, which blocks glycine receptors (GlyRs), suggesting that the abnormal motor response in mutants may be attributable to a deficit in glycinergic synaptic transmission. We identified a missense mutation in the gene encoding RNA helicase Dhx37. In Dhx37 mutants, GlyR alpha subunit mRNA levels were decreased due to a splicing defect. Overexpression of GlyR alpha subunits in Dhx37-deficient embryos restored normal behavior. Conversely, antisense knockdown of multiple GlyR alpha subunits in wild-type embryos was required to recapitulate the Dhx37 mutant phenotype. These results indicate that Dhx37 is specifically required for the biogenesis of a subset of GlyR alpha subunit mRNAs, thereby regulating glycinergic synaptic transmission and associated motor behaviors.
A way of making tamed animals from wild mice
Mouse Genomics Resource Laboratory (MGRL) • Koide Group
Tatsuhiko Goto, Akira Tanave, Kazuo Moriwaki, Toshihiko Shiroishi, Tsuyoshi Koide
Genes Brain and Behavior 2013. DOI: 10.1111/gbb.12088
Humans have developed many domestic animals for various purposes. Tameness is a behavioral characteristic that is changed during domestication process of wild ancestors. However, it is still need to be clarified how tameness change the behavior of animals. In this point of view, we took particular note of a book written by an American zoologist, Edward O. Price. He defined tameness as “a measure of the extent to which an individual is reluctant to avoid or motivated to approach humans.” In order to address this point, we developed three behavioral tests to measure the level of two different classes of tameness in mice. We characterized tame behavior using 17 inbred mouse strains: ten wild strains, one Japanese fancy-mouse strain, and six laboratory strains. As a results, most of the domesticated strains showed significantly greater reluctance to avoid humans than wild strains, whereas there was no significant difference in the level of motivation to approach humans between these two groups. These results suggest that domesticated strains were predominantly selected for reluctance to avoid humans over the course of their domestication history.
Dr. Tatsuhiko Goto worked on this project as a project researcher in Transdisciplinary Research Integration Center.
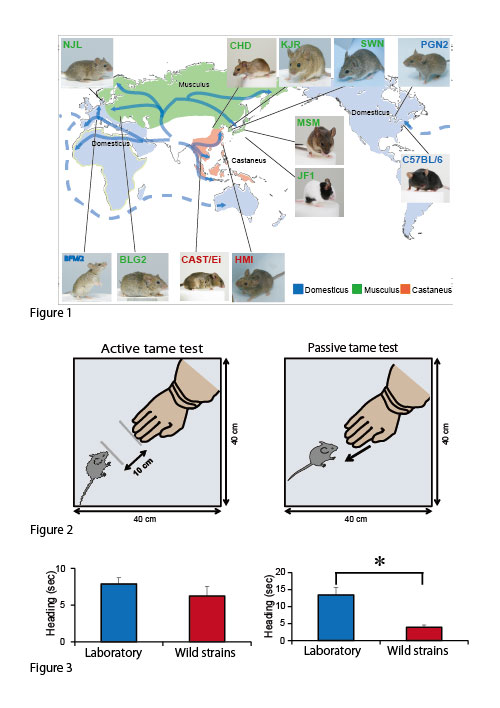
Figure 1. Dr. Kazuo Moriwaki, an emeritus professor, introduced wild mice from many countries and developed wild-derived inbred strains (wild strains). Given that these strains have not been subjected to deliberate attempts at domestication during inbreeding, these mice still show the characteristic behavior of wildness, such as quick movements, aggression, and higher responsiveness to handling by humans.
Figure 2. Tame tests developed for measuring levels of tameness in mice
Figure 3. Most of the domesticated strains showed significantly higher level of reluctance to avoid human than wild-derived mice, whereas there was no significant difference in the level of motivated to approach human.
Rings in response to light cycles
Microbial Genetics Laboratory • Niki Group
Sho Okamoto, Kanji Furuya, Shingo Nozaki, Keita Aoki, and Hironori Niki
Eukaryotic Cell 2013 12:1235-1243. doi:10.1128/EC.00109-13
Many fungi respond to light and regulate fungal development and behavior. A blue light-activated complex has been identified in Neurospora crassa as the product of the wc-1 and wc-2 genes. Orthologs of WC-1 and WC-2 have hitherto been found only in filamentous fungi and not in yeast, with the exception of the basidiomycete pathogenic yeast Cryptococcus. Here, we report that the fission yeast Schizosaccharomyces japonicus responds to blue light depending on Wcs1 and Wcs2, orthologs of components of the WC complex. Surprisingly, those of ascomycete S. japonicus are more closely related to those of the basidiomycete. S. japonicus reversibly changes from yeast to hyphae in response to environmental stresses. After incubation at 30°C, a colony of yeast was formed, and then hyphal cells extended from the periphery of the colony. When light cycles were applied, distinct dark- and bright-colored hyphal cell stripes were formed because the growing hyphal cells had synchronously activated cytokinesis. In addition, temperature cycles of 30°C for 12 h and 35°C for 12 h or of 25°C for 12 h and 30°C for 12 h during incubation in the dark induced a response in the hyphal cells similar to that of light. The stripe formation of the temperature cycles was independent of the wcs genes. Both light and temperature, which are daily external cues, have the same effect on growing hyphal cells. A dual sensing mechanism of external cues allows organisms to adapt to daily changes of environmental alteration.
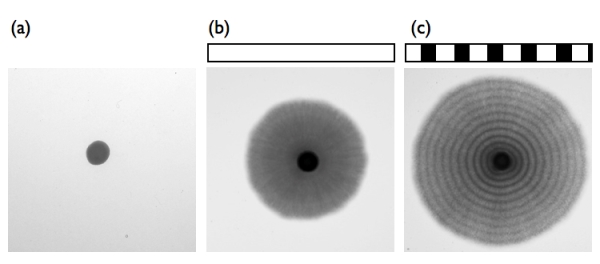
After incubation at 30°C, a colony of yeast was formed (a), and then hyphal cells extended from the periphery of the colony (b). When light cycles were applied, distinct dark- and bright-colored hyphal cell stripes were formed because the growing hyphal cells had synchronously activated cytokinesis (c).
Muscle disease gene Stac3 identified in fish
Motor Neural Circuit Laboratory • Hirata Group
Horstick, E. J., Linsley, J. W., Dowling, J. J., Hauser, M. A., McDonald, K. K., Ashley-Koch, A., Saint-Amant, L., Satish, A., Cui, W. W., Zhou, W., Sprague, S. M., Stamm, D. S., Powell, C. M., Speer, M. C., Franzini-Armstrong, C., Hirata, H.* and Kuwada, J. Y.* (*Corresponding authors)
Nature Communications 4: 1952 (2013). doi:10.1038/ncomms2952
Native American myopathy (NAM) is a congenital muscular disease prevalent in the southern regions of the United States. However, the cause of this intractable myopathy has not been clarified so far. We originally studied mutant zebrafish that exhibited severe muscle weakness. The responsible gene encoded for a muscle protein Stac3. We found that Stac3 associates with dihydropyridine and ryanodine receptors and regulates calcium release during muscle contraction. Stac3-deficient zebrafish is now used for identifying drugs that mitigate the muscular defect in humans.
This is a collaborative work with Dr. John Y. Kuwada (University of Michigan).
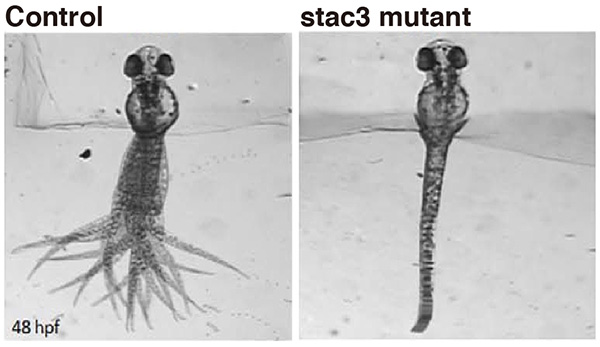
Touch evoked swimming in wild-type (control) but not stac3 mutant zebrafish embryos at 48 hours post-fertilization. Panels show superimposed frames of swimming motion with the head embedded in agarose.
ZC4H2 mutations are associated with arthrogryposis multiplex congenita and intellectual disability
Motor Neural Circuit Laboratory • Hirata Group
Hirata, H.*, Nanda, I.*, van Riesen, A.*, McMichael, G.*, Hu, H.*, Hambrock, M., Papon, M.-A., Fischer, U., Marouillat, S., Ding, C., Alirol, S., Bienek, M., Preisler-Adams, S., Grimme, A., Seelow, D., Webster, R., Haan, E., MacLennan, A., Stenzel, W., Yap, T. Y., Gardner, A., Nguyen, L. S., Shaw, M., Lebrun, N., Haas, S. A., Kress, W., Haaf, T., Schellenberger, E., Chelly, J., Viot, G., Shaffer, L. G., Rosenfeld, J. A., Kramer, N., Falk, R., El-Khechen, D., Escobar, L. F., Hennekam, R., Wieacker, P., Hübner, C., Ropers, H.-H., Gecz, J., Schuelke, M., Laumonnier, F. and Kalscheuer, V. M. (*Equal contribution)
American Journal of Human Genetics 92: 681-695 (2013). doi: 10.1016/j.bbr.2011.03.031
Arthrogryposis multiplex congenita (AMC) is caused by heterogeneous pathologies leading to multiple antenatal joint contractures through fetal akinesia. Understanding the pathophysiology of this disorder is important for clinical care of the affected individuals and genetic counseling of the families. In this study, we identified disease-causing mutations in the zinc-finger gene ZC4H2 in an AMC subtype that is associated with multiple dysmorphic features and intellectual disability (ID). In zebrafish, antisense-morpholino-mediated zc4h2 knockdown caused abnormal swimming and impaired primary motoneuron development. All missense mutations identified herein failed to rescue the swimming defect of zebrafish morphants. We conclude that ZC4H2 defects cause a clinically variable broad-spectrum neurodevelopmental disorder of the central and peripheral nervous systems. Our results highlight the importance of ZC4H2 for genetic testing of individuals presenting with ID plus muscle weakness and minor or major forms of AMC.
This is a collaborative work with Dr. Vera M. Kalscheuer (Max Planck Institute for Molecular Genetics), Dr. Markus Schuelke (Charité Universitätsmedizin Berlin) and other researchers and is supported by the NIG Collaborative Research.
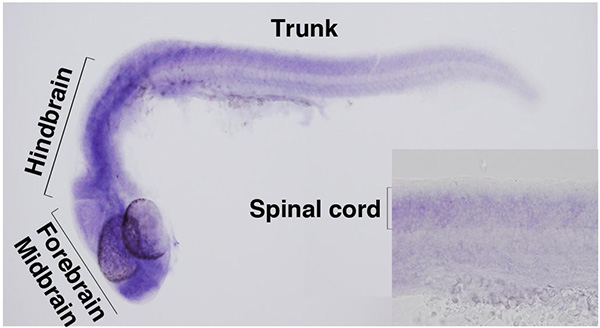
In zebrafish embryos, zc4h2 is expressed by forebrain, midbrain, hindbrain and spinal cord.
Mechanism for transcribing Arabidopsis genes containing intragenic heterochromatin
Division of Agricultural Genetics • Kakutani Group
Hidetoshi Saze, Junko Kitayama, Kazuya Takashima, Saori Miura, Yoshiko Harukawa, Tasuku Ito and Tetsuji Kakutani
Nature Communications, 4, Article number:2301 doi:10.1038/ncomms3301
Genomes of vertebrates and plants contain a substantial number of transposable elements (TEs), which are silenced by repressive epigenetic modifications, such as cytosine methylation and methylation of lysine 9 of histone H3. These modifications are essential for formation of inactive chromatin structures called heterochromatin. A potential complication is that active cellular genes sometimes contain TEs within their transcribed regions.
In this study, we show that heterochromatic epigenetic modifications are commonly found within actively transcribed gene units in both the Arabidopsis and rice genomes. We further show that in Arabidopsis, full-length transcription of genes with intragenic heterochromatin, most of which is formed by TE insertions, requires IBM2 (Increase in Bonsai Methylation 2), a protein with a Bromo-Adjacent Homology (BAH) domain and an RNA recognition motif (RRM). Our results reveal a novel epigenetic mechanism that masks effects of genetic variations created by TE insertions, allowing evolution of complex genomes with heterochromatic domains having diverse functions. (This work is a collaboration with Saze lab in OIST.)
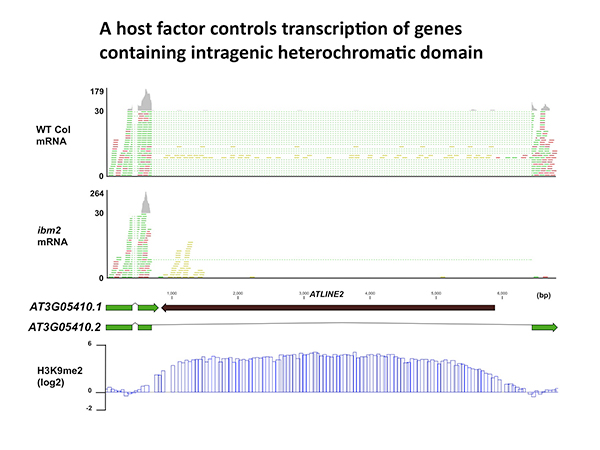
Full-length transcription of the gene containing heterochromatin is impaired in ibm2 mutant. mRNA reads are indicated with green and red. Reads with dotted lines are mapped across exon-exon boundaries. Note that the inserted TE (ATLINE2) is associated with heterochromatic H3K9 methylation (bottom).
Interhemispheric asymmetry of olfactory input-dependent neuronal specification in the adult zebrafish brain
Division of Molecular and Developmental Biology • Kawakami Group
Kishimoto, N., Asakawa, K., Madelaine, R., Blader, P., Kawakami, K., and Sawamoto, K.
Nature Neuroscience 16, 884-888, 2013 doi:10.1038/nn.3409
The vertebrate brain is anatomically and functionally asymmetric. The left and right cerebral hemispheres harbor neural stem cell niches at the ventricular-subventricular zone (V-SVZ) of the ventricular walls, where new neurons are continuously generated throughout life. However, any interhemispheric asymmetry of neural stem cell niches remains unclear. We performed gene-trap screens in adult zebrafish to identify genes that are differentially expressed in the two hemispheres and found that adult-born neurons expressing the neural zinc-finger protein Myt1 exist predominantly in the left V-SVZ. This lateralization could be reversed by left olfactory sensory deprivation–induced inactivation of Notch signaling. The olfactory behavioral preference for attractive amino acids was also impaired by sensory deprivation of the left olfactory system, but not of the right olfactory system. Our findings suggest that olfactory input generates interhemispheric differences in the fate of adult-born neurons in the zebrafish brain.
This study has been carried out as collaboration with Drs. Kishimoto and Sawamoto at Nagoya City University.
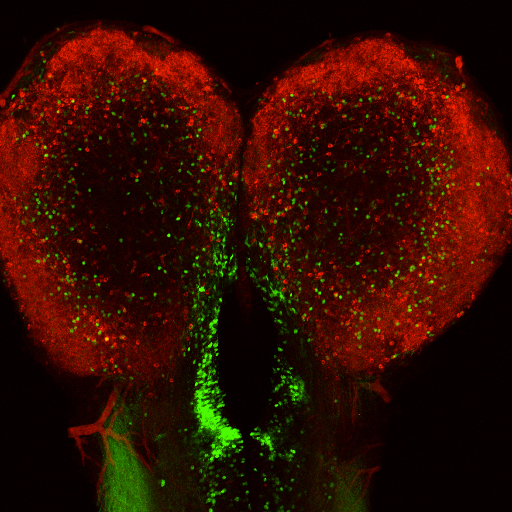
Interhemispheric asymmetric distribution of Myt1-positive neurons in telencephalic ventricular-subventricular zone.
Blood flow-dependent expression of miR-21 controls valvulogenesis of the zebrafish heart
Division of Molecular and Developmental Biology • Kawakami Group
Toshihiro Banjo,Janin Grajcarek,Daisuke Yoshino,Hideto Osada,Kota Y. Miyasaka,Yasuyuki S. Kida,Yosuke Ueki,Kazuaki Nagayama,Koichi Kawakami,Takeo Matsumoto,Masaaki Sato & Toshihiko Ogura
Nature Communications 4,Article number:1978,2013 doi:10.1038/ncomms2978
Heartbeat is required for normal development of the heart, and perturbation of intracardiac flow leads to morphological defects resembling congenital heart diseases. These observations implicate intracardiac haemodynamics in cardiogenesis, but the signalling cascades connecting physical forces, gene expression and morphogenesis are largely unknown. Here we use a zebrafish model to show that the microRNA, miR-21, is crucial for regulation of heart valve formation. Expression of miR-21 is rapidly switched on and off by blood flow. Vasoconstriction and increasing shear stress induce ectopic expression of miR-21 in the head vasculature and heart. Flow-dependent expression of mir-21 governs valvulogenesis by regulating the expression of the same targets as mouse/human miR-21 (sprouty, pdcd4, ptenb) and induces cell proliferation in the valve-forming endocardium at constrictions in the heart tube where shear stress is highest. We conclude that miR-21 is a central component of a flow-controlled mechanotransduction system in a physicogenetic regulatory loop.
This study has been carried out as collaboration with Dr. Ogura at Tohoku University.
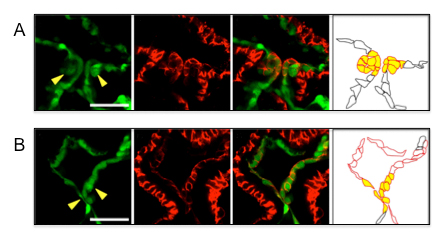
A: Normal valvulogenesis in the wild type zebrafish embryo.
B: Abnormal valvulogenesis in the zebrafish embryo in which the function of miR-21 was inhibited.
Recovery of rod-shaped cells by delayed cell division
Microbial Genetics Laboratory • Niki Group
Daisuke Shiomi, Hironori Niki
MicrobiologyOpen (DOI: 10.1002/mbo3.116)
RodZ is important for maintaining the rod shape of Escherichia coli. Loss of RodZ causes conversion of the rod shape to a round shape and a growth rate slower than that of wild-type cells. Suppressor mutations that simultaneously restore both the growth rates and the rod shape were isolated. Most of the suppressor mutations are found in mreB, mrdA, or mrdB. One of the mutations was in the promoter region of zipA, which encodes a crucial component of the cell division machinery. In this study, we investigated the mechanism of the suppression by this mutation. ZipA was slightly but significantly increased in the suppressor cells and led to a delay in cell division. While round-shaped mreB and mrdA mutants lose cell bipolarity, we found that round-shaped rodZ mutants retained cell bipolarity. Therefore, we concluded that a delay in the completion of septation provides extra time to elongate the cell laterally so that the zipA suppressor mutant is able to recover its ovoid or rod shape. The suppression by zipA demonstrates that the regulation of timing of septation potentially contributes to the conversion of morphology in bacterial cells.
This study has been carried out as collaboration with Dr. Ogura at Tohoku University.
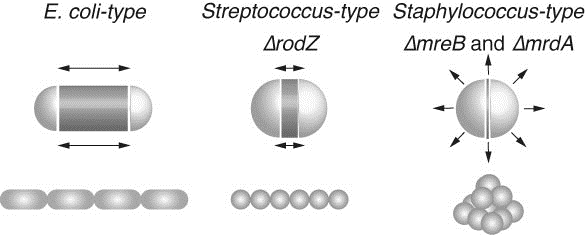
Rod shaped E. coli cells grow at the central cylinder. Ovoid shaped Streptococcus cells grow at septum while round shaped Staphylococcus cells swell. Black arrows indicate the direction of increase of cell volume. The increase in cell volume of ΔrodZ resembles that of Streptococcus, while those of ΔmreB or ΔmrdA resemble that of Staphylococcus. Dark gray zones indicate regions where peptidoglycan is actively synthesized. E. coli (WT), ΔrodZ, and Streptococcus cells retain cell polarity, while E. coli ΔmreB, ΔmrdA, and Staphylococcus cells lose polarity.
Chromosome condensation is affected by the size of the cell nucleus
Cell Architecture Laboratory • Kimura Group
Yuki Hara, Mari Iwabuchi, Keita Ohsumi, Akatsuki Kimura
Mol. Biol. Cell August 1, 2013 vol. 24(15) 2442-2453 doi: 10.1091/mbc.E13-01-0043
Chromosome condensation is critical for accurate inheritance of genetic information. The degree of condensation, which is reflected in the size of the condensed chromosomes during mitosis, is not constant. It is differentially regulated in embryonic and somatic cells. In addition to the developmentally programmed regulation of chromosome condensation, there may be adaptive regulation based on spatial parameters such as genomic length or cell size. We propose that chromosome condensation is affected by a spatial parameter called the chromosome amount per nuclear space, or “intranuclear DNA density.” Using Caenorhabditis elegans embryos, we show that condensed chromosome sizes vary during early embryogenesis. Of importance, changing DNA content to haploid or polyploid changes the condensed chromosome size, even at the same developmental stage. Condensed chromosome size correlates with interphase nuclear size. Finally, a reduction in nuclear size in a cell-free system from Xenopus laevis eggs resulted in reduced condensed chromosome sizes. These data support the hypothesis that intranuclear DNA density regulates chromosome condensation. This suggests an adaptive mode of chromosome condensation regulation in metazoans.
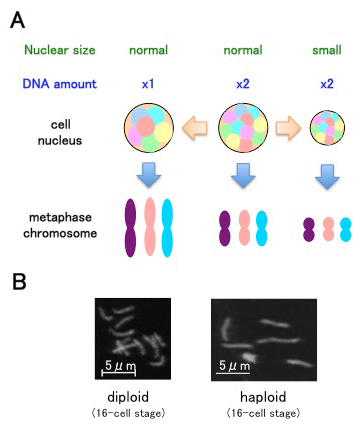
(A) Intranuclear DNA density affects chromosome condensation. When the nucleus is smaller, metaphase chromosomes are shorter. When the nucleus contains less chromosomal DNA, metaphase chromosomes are longer.
(B) Examples of metaphase chromosomes from C. elegans samples.
Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor
Division of Agricultural Genetics • Kakutani Group
Yu Fu, Akira Kawabe, Mathilde Etcheverry, Tasuku Ito, Atsushi Toyoda, Asao Fujiyama, Vincent Colot, Yoshiaki Tarutani, Tetsuji Kakutani
EMBO Journal, advance online publication 30 July 2013; doi:10.1038/emboj.2013.169
Transposable elements (TEs) have a major impact on genome evolution, but they are potentially deleterious, and most of them are silenced by epigenetic mechanisms such as DNA methylation. Here we report the characterization of a TE encoding an activity to counteract epigenetic silencing by the host. In Arabidopsis thaliana, we identified a mobile copy of the Mutator-like element (MULE) with degenerated terminal inverted repeats (TIRs). This TE, named Hiun (Hi), is silent in wild type plants, but it transposes when DNA methylation is abolished. When a Hi transgene was introduced into the wild type background, it induced excision of the endogenous Hi copy, suggesting that Hi is the autonomously mobile copy. In addition, the transgene induced loss of DNA methylation and transcriptional activation of the endogenous Hi. Most importantly, the trans-activation of Hi depends on a Hi-encoded protein different from the conserved transposase. Proteins related to this anti-silencing factor, which we named VANC, are widespread in the non-TIR MULEs and may have contributed to the recent success of these TEs in natural Arabidopsis populations.
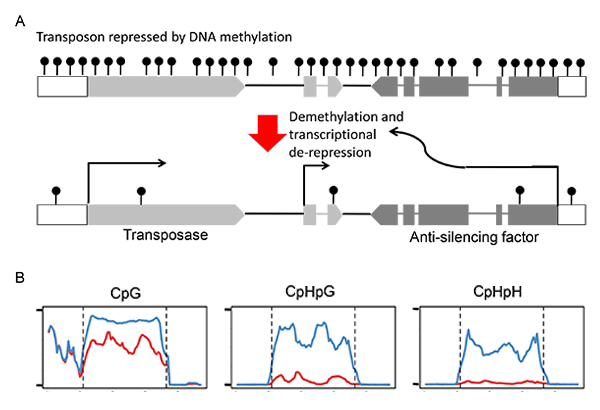
(A) Structure of the plant transposon Hiun (Hi). This transposon encodes for the transposase and the anti-silencing factor.
The antisilencing factor induces demethylation and transcriptional de-repression.
(B) DNA methylation of endogenous Hi compared between non-transgenic plant (blue) and transgenic plant expressing Hi transgene (red).
The transgene induced demethylaton of endogenous copy, especially at non-CpG sites.
Wnt/Dkk negative feedback regulates sensory organ size in zebrafish
Division of Molecular and Developmental Biology • Kawakami Group
H. Wada, A. Ghysen, K. Asakawa, G. Abe, T. Ishitani, K. Kawakami.
Current Biology, 23 (16), p1559–1565, DOI: 10.1016/j.cub.2013.06.035
Correct organ size must involve a balance between promotion and inhibition of cell proliferation. A mathematical model has been proposed, where an organ is assumed to produce its own growth activator, as well as a growth inhibitor. But, there is yet no molecular evidence to support this model. The mechanosensory organs of the fish lateral line system (neuromasts) are composed of a core of sensory hair cells surrounded by non-sensory support cells. Sensory cells are constantly replaced, and are regenerated by surrounding non-sensory cell, while each organ retains the same size throughout life. Moreover, neuromasts also bud off new neuromasts, which stop growing when they reach the same size. In this study, we show that the size of neuromasts is controlled by a balance between growth-promoting Wnt signaling activity in proliferation-competent cells, and Wnt-inhibiting Dkk activity produced by differentiated sensory cells. This negative feedback loop from Dkk (secreted by differentiated cells) on Wnt-dependent cell proliferation (in surrounding cells) also acts during regeneration to achieve size constancy. This study establishes Wnt/Dkk as a novel mechanism to determine the final size of an organ.
This study was funded by the PRESTO of the Japan Science and Technology Agency.

A) Schematic drawing of a neuromast.
(B) Schematic representation of Wnt signaling and of its inhibition by Dkk signaling.
(C) Wnt reporter activity (green) gradually subsides as hair cells (red) are formed.
(D) Dkk2 expression coincides with neuromast maturation.
Detailed analysis of a complex copy number variation region in mouse genome
Mouse Genomics Resource Laboratory (MGRL) • Koide Group
Umemori J, Mori A, Ichiyanagi K, Uno T, and Koide T.
BMC Genomics14: 455, 2013.
Copy number variation (CNV) of genomic segments is a common phenomenon that affects more than 10% of the human and mouse genomes, and is an important source of diversity in genomic structure. CNVs are frequently found in clusters called CNV regions (CNVRs), which are strongly associated with large segmental duplications (large SDs). However, the composition of these complex repetitive structures remains unclear. In the present study, we established new method for analyzing on complex repetitive structures of CNVRs by collaborating with National Institute of Informatics.
At first, we conducted self-comparative-plot analysis of all mouse chromosomes using the high-speed and large-scale-homology search algorithm, Similarity/Homology Efficient Analyze Procedure (SHEAP) developed by Dr. Takeaki Uno in National Institute of Informatics. By using this method, large SDs were visualized as unique tartan-checked patterns with complex arrangement of diagonal split lines (Figure 1). We focused on one SD on chromosome 13 (SD13M), which is one of the causative regions for genetic incompatibility (papers in preparation), and applied blast-based Systematic analysis of HErPlot to Extract Regional Distinction (SHEPHERD), a stepwise ab initio method, to extract core elements of repetitive sequences in SD13M (Figure 2). Then, comparative genome hybridization array analysis was empirically conducted on MSM, BLG2 (strains derived from wild mice in Mishima in Japan, and Toshevo in Bulgaria, respectively), and C57BL/6J (an experimental strain). This analysis showed that core elements are categorized to ones with CNVs and the others with constant copy number among strains, which have distinctive characters and divergences. The present study seems to be helpful for elucidating evolutional processes and functions of the CNVRs (Fig. 3).
This study was funded by the “Cultivation of integrated project” of Transdisciplinary Research Integration Center in Research Organization of Informatio and Systems (Japan).
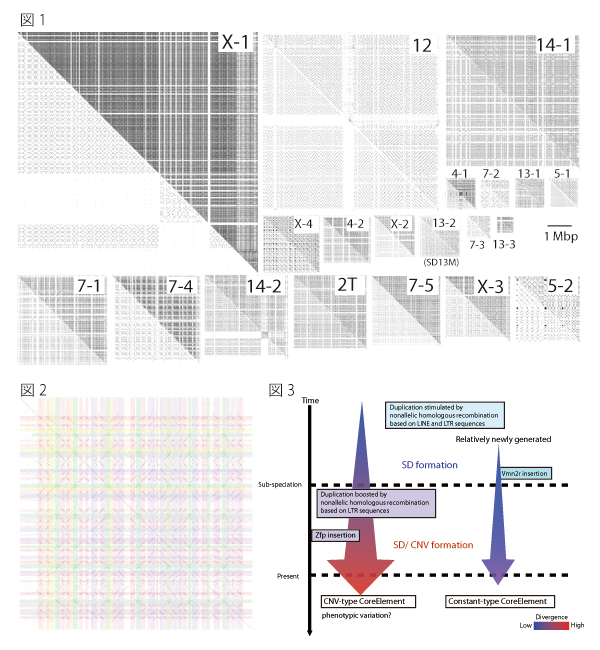
Fig.1. Tartan-checked structure of SDs visualized using SHEAP.
The lower left triangle of each panel shows a self-plot of the sequence after known repeat sequences have been masked using RepeatMasker. Each of the upper right triangles shows a self-plot of the intact sequence. Diagonal lines aligned in the same column or row represent repetitive sequences.
Fig.2. Extraction of repeat units from the self-plot of the large SD
Diagonal lines were extracted from a self-comparative-plot of SD13M that consisted of a dot-plot matrix. Then we selected one sequence and removed the other sequences represented by diagonal lines that were located in the same column or in the same row.
Fig.3 Model for the formation of SDs and CNVs
The average divergence of CNV-type core elements was greater than that of the constant type, and the CNV-type core elements contained significantly larger proportions of long terminal repeat (LTR) types of retrotransposon than the constant-type core elements. These results suggest that constant-type core elements emerged more recently than CNV-type core elements, and that retrotransposition of LTRs promotes nonallelic homologous recombination and caused CNV in SD13M.
Concerted interaction between origin recognition complex (ORC), nucleosomes, and replication origin DNA ensures stable ORC–origin binding
Division of Microbial Genetics • Araki Group
Kohji Hizume, Masaru Yagura, and Hiroyuki Araki
Genes to Cells , 24 JUN 2013 DOI: 10.1111/gtc.12073. [Epub ahead of print]
Chromosomal replication origins, where DNA replication is initiated, are determined in eukaryotic cells by specific binding of a six-subunit origin recognition complex (ORC). Many biochemical analyses have revealed the detailed properties of the ORC–DNA interaction. However, because of the lack of in vitro analysis, the molecular architecture of the ORC–chromatin interaction is unclear. Recently, mainly from in vivo analyses, a role of chromatin in the ORC–origin interaction has been reported, including the existence of a specific pattern of nucleosome positioning around origins and of a specific interaction between chromatin—or core histones—and Orc1, a subunit of ORC. Therefore, to understand how ORC establishes its interaction with origin in vivo, it is essential to know the molecular mechanisms of the ORC–chromatin interaction. Here, we show that ORC purified from yeast binds more stably to origin-containing reconstituted chromatin than to naked DNA, and forms a nucleosome-free region at origins. Molecular imaging using atomic force microscopy (AFM) reveals that ORC associates with the adjacent nucleosomes and forms a larger complex. Moreover, stable binding of ORC to chromatin requires linker DNA. Thus, ORC establishes its interaction with origin by binding to both nucleosome-free origin DNA and neighboring nucleosomes.
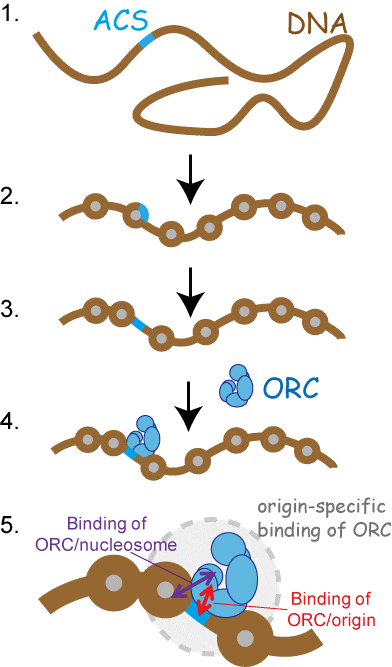
(1) DNA fragment containing origin-specific DNA motif (ACS).
(2) By chromatin reconstitution, nucleosome is formed on ACS.
(3) By the addition of ORC, ACS becomes nucleosome-free-region.
(4) ORC interacted to nucleosome-free ACS.
(5) ORC establishes its interaction with origin by binding to both nucleosome-free ACS and neighboring nucleosomes.
Regulation of nuclear envelope dynamics that is necessary for the progression of semi-open mitosis
Microbial Genetics Laboratory • Niki Group
Keita Aoki, Yuh Shiwa, Hiraku Takada, Hirofumi Yoshikawa and Hironori Niki
Genes to Cells 21 JUN 2013 DOI: 10.1111/gtc.12072. [Epub ahead of print]
Three types of mitosis, which are open, closed, or semi-open mitosis, function in eukaryotic cells, respectively. The open mitosis involves breakage of the nuclear envelope before nuclear division, whereas the closed mitosis proceeds with an intact nuclear envelope. To understand the mechanism and significance of three types of mitotic division in eukaryotes, we investigated the process of semi-open mitosis, in which the nuclear envelope is only partially broken, in the fission yeast Schizosaccharomyces japonicus. In anaphase-promoting complex/cyclosome (APC/C) mutants of Sz. japonicus, the nuclear envelope remained relatively intact during anaphase, resulting in impaired semi-open mitosis. As a suppressor of apc2 mutant, a mutation of Oar2 which was a 3-oxoacyl-[acyl-carrier-protein] reductase was obtained. The level of the Oar2, which had two destruction-box motifs recognized by APC/C, was increased in APC/C mutants. Furthermore, the defective semi-open mitosis observed in an apc2 mutant was restored by mutated oar2+. Based on these findings, we propose that APC/C regulates the dynamics of the nuclear envelope through degradation of Oar2 dependent on APC/C during the metaphase-to-anaphase transition of semi-open mitosis in Sz. japonicus.
This study has been carried out as collaboration with Dr. Ogura at Tohoku University.
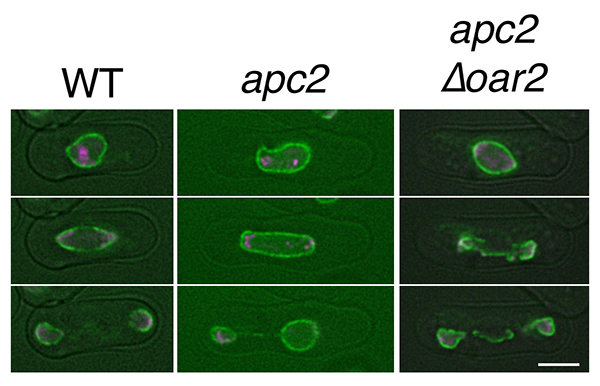
Observations of nuclear envelope dynamics in WT, apc2 mutant, and apc2∆oar2 mutant. The semi-open mitosis appeared in WT was defective in apc2 mutant, but partially restored in apc2∆oar2 mutant. Green: nuclear envelope marker (Cut11-GFP), magenta: chromosomal marker (H2A-mCherry). Scale bar: 5µm.
An allometric relationship between mitotic spindle width, spindle length, and ploidy in Caenorhabditis elegans embryos
Cell Architecture Laboratory • Kimura Group
Yuki Hara, Akatsuki Kimura
Mol. Biol. Cell 24 (9), 1411-1419, 2013 doi:10.1091/mbc.E12-07-0528
The mitotic spindle is a diamond-shaped molecular apparatus crucial for chromosomal segregation. Previous studies suggested that the spindle can self-organize to maintain a constant aspect ratio between its length and width against physical perturbations. Here we determine the widths of metaphase spindles of various sizes observed during embryogenesis in Caenorhabditis elegans. The spindle width correlates well with the spindle length, but the aspect ratio between the spindle length and spindle width is not constant, indicating an allometric relationship between these parameters. We characterize how DNA quantity (ploidy) affects spindle shape by using haploid and polyploid embryos. On the basis of the quantitative data, we deduce an allometric equation that describes the spindle width as a function of the length of the hypotenuse and ploidy. On the basis of this equation, we propose a force-balance model to determine the spindle width.
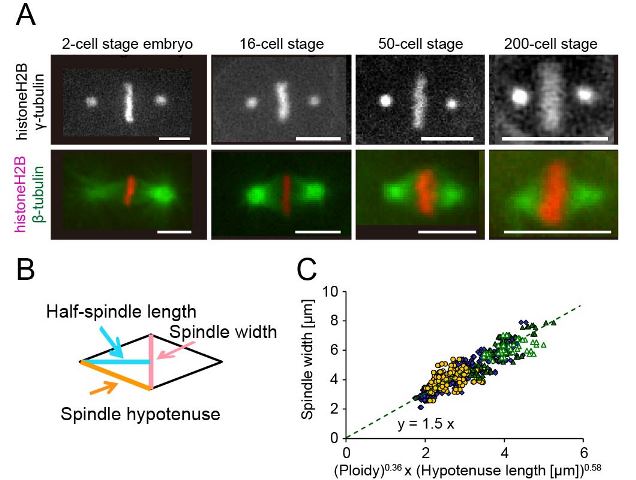
(A) Metaphase spindles at 4 representative cell stages (Bar = 5 μm). (B) The spindle width (pink), half-spindle length (light blue), and hypotenuse length (orange) were shown. (C) The spindle width from diploid (blue diamond), haploid (yellow circle), and polyploid (green triangles) is plotted against the product of 0.36th-power of ploidy and 0.58th-power of hypotenuse length. (Figures were modified from the paper.)















