Niki Group • Microbial Physiology Laboratory
Genetic dissection of the cell division mechanism using single-cellular model organisms
Faculty
Research Summary
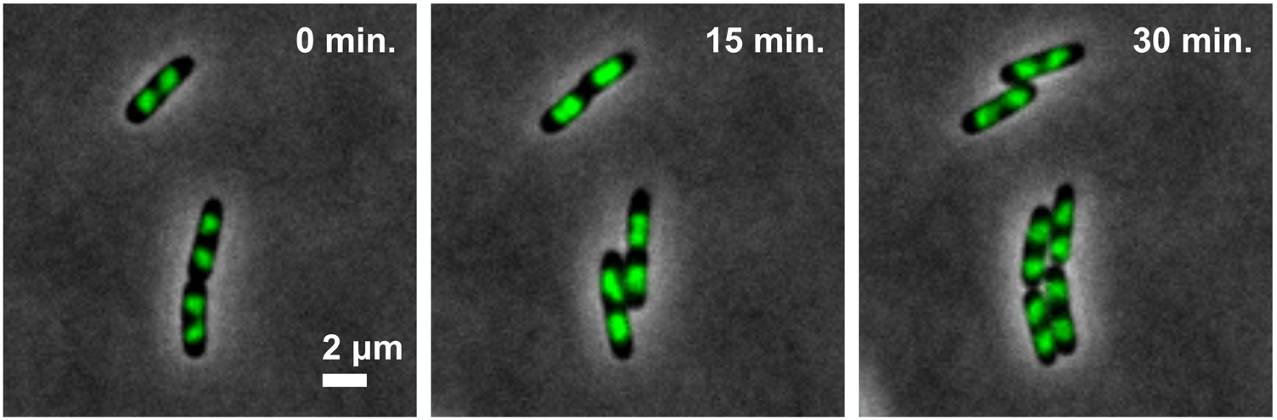
Bacteria and yeast are important model organisms to elucidate the fundamental mechanisms of cell proliferation. Our laboratory studies the mechanisms behind the cell division cycle and adaptations to external stresses under environments. We focused on compaction of chromosomal DNA as a nucleoid inside a tiny bacterial cell during cell division. Bacterial condensin is an essential factor for packaging of a nucleoid to properly segregate into daughter cells. Also, we study on hyphal development and growth by using a new model organism, Schizosaccharomyces japonicus. We established new investigative methodologies to investigated S. japonicus.
https://shigen.nig.ac.jp/ecoli/strain/
https://shigen.nig.ac.jp/bsub/
Selected Publications
Fujiwara K, Tsuji N, Yoshida M, Takada H, Chiba S. Patchy and widespread distribution of bacterial translation arrest peptides associated with the protein localization machinery. Nat Commun. 2024 Apr 2;15(1):2711.
Yano K, Noguchi H, Niki H. Profiling a single-stranded DNA region within an rDNA segment that affects the loading of bacterial condensin. iScience. 2022 Nov 4;25(12):105504.
Seike T, Sakata N, Shimoda C, Niki H, Furusawa C. The sixth transmembrane region of a pheromone G-protein coupled receptor, Map3, is implicated in discrimination of closely related pheromones in Schizosaccharomyces pombe. Genetics. 2021 Dec 10;219(4):iyab150.
Nakai R, Wakana I, Niki H. Internal microbial zonation during the massive growth of marimo, a lake ball of Aegagropila linnaei in Lake Akan. iScience. 2021 Jun 12;24(7):102720.

















