Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Gen Shiratsuchi and Tadasu Nozaki were awarded poster prize at the 65th Annual Meeting of the Japan Society for Cell Biology.
 The 65th Annual Meeting of the Japan Society for Cell Biology
The 65th Annual Meeting of the Japan Society for Cell Biology
 Kitagawa Laboratory (Center for Frontier Research)
Kitagawa Laboratory (Center for Frontier Research)
 Maeshima Laboratory (Structure Biology Center)
Maeshima Laboratory (Structure Biology Center) Efficient Initiation of DNA Replication in Eukaryotes Requires Dpb11/ TopBP1-GINS Interaction.
Division of Microbial Genetics • Araki Group
Seiji Tanaka, Yayoi Komeda, Toshiko Umemori, Yumiko Kubota, Haruhiko Takisawa, and Hiroyuki Araki
Mol. Cell. Biol.(2013), 33 2614-2622. doi:10.1128/MCB.00431-13
Dpb11/Cut5/TopBP1 is evolutionarily conserved and is essential for the initiation of DNA replication in eukaryotes. The Dpb11 of the budding yeast Saccharomyces cerevisiae has four BRCT domains (BRCT1 to -4). The N-terminal pair (BRCT1 and -2) and the C-terminal pair (BRCT3 and -4) bind to cyclin-dependent kinase (CDK)-phosphorylated Sld3 and Sld2, respectively. These phosphorylation-dependent interactions trigger the initiation of DNA replication. BRCT1 and -2 and BRCT3 and -4 of Dpb11 are separated by a short stretch of ~100 amino acids. It is unknown whether this inter-BRCT region functions in DNA replication. Here, we showed that the inter-BRCT region is a GINS interaction domain that is essential for cell growth and that mutations in this domain cause replication defects in budding yeast. We found the corresponding region in the vertebrate ortholog, TopBP1, and showed that the corresponding region also interacts with GINS and is required for efficient DNA replication. We propose that the inter-BRCT region of Dpb11 is a functionally conserved GINS interaction domain that is important for the initiation of DNA replication in eukaryotes.
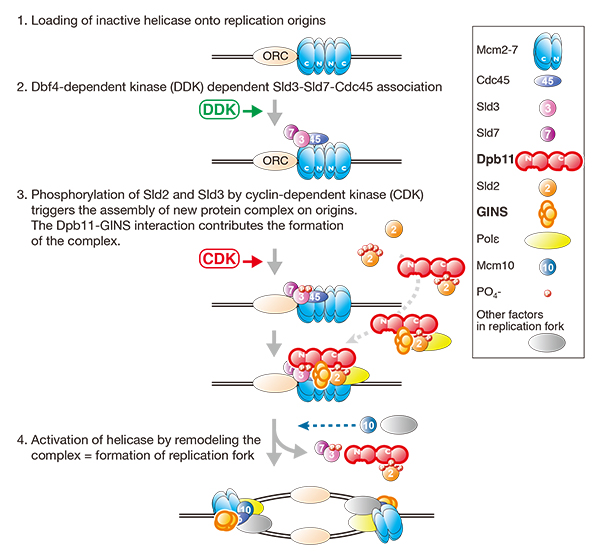
Schematic drawings of the initiation of DNA replication in eukaryotes. Dpb11-GINS interaction is important for the assembly of protein complex, which is required for the activation of replicative helicase.
See-through motor column
Division of Molecular and Developmental Biology・Kawakami Group
Kazuhide Asakawa, Gembu Abe, and Kawakami Koichi
Frontiers in Neural Circuits, 2013 May 28;7:100. DOI: 10.3389/fncir.2013.00100
The spatio-temporal regulation of muscle contractions that generate body movements critically depends on the exquisitely precise innervation of each muscle type by the appropriate motoneuron subtype. Therefore, delineating the identities of motoneurons and their connectivity to target muscles is fundamental to an understanding of the motor control by the central nervous system. In this study, by taking advantage of the optical and genetic accessibility, we dissected the spinal cord motor column of zebrafish larvae at the cellular level. By using the BAC for the Mnx homeodomain gene mnr2b, we established the mnGFF7 transgenic line expressing the Gal4FF transcription factor in a large part of the motor column. Single cell labelling of Gal4FF-expressing cells in the mnGFF7 larvae enabled a detailed investigation of the morphological characteristics of individual spinal motoneurons, as well as the overall organisation of the motor column in a spinal segment. The transgenic fish established here should facilitate an understanding of the cellular and molecular architecture of the spinal cord motor column and its connection to muscles in vertebrates.
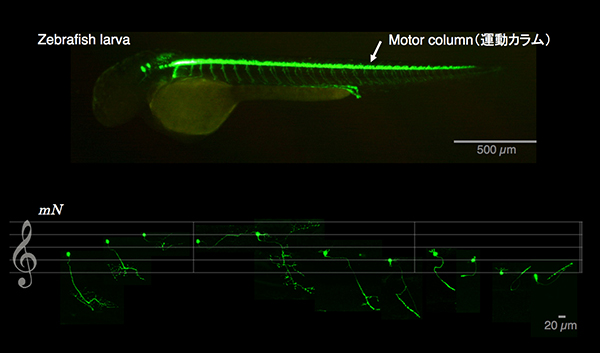
Lateral view of a zebrafish larva (top). Eleven different types of spinal motoneurons identified in the single cell labeling experiment.
The ancestor of extant Japanese fancy mice contributed to the mosaic genomes of classical inbred strains.
Press release
The ancestor of extant Japanese fancy mice contributed to the mosaic genomes of classical inbred strains
Takada, T., Ebata, T., Noguchi, H., Keane, K., Adams, D., Narita, T., Shin-I, T., Fujisawa, H., Toyoda, A., Abe, K., Obata, Y., Sakaki, Y., Moriwaki, K., Fujiyama, A., Kohara, Y. and Shiroishi, T.Genome Research, 2013 Aug;23(8):1329-1338. DOI: 10.1101/gr.156497.113
We resequenced the genomes of Mus musculus molossinus-derived two inbred strains, MSM/Ms and JF1/Ms. MSM/Ms originated from Japanese wild mice, and ancestry of JF1/Ms was originally found in Europe and then transferred to Japan. We compared the characteristics of these sequences to those of the C57BL/6J reference sequence and the recent datasets from the resequencing of 17 inbred strains in the Mouse Genome Project.
The major outcomes of this study are summarized below.
1. Over 10 million SNPs and 1 million short indels are identified between the MSM/Ms or JF1/Ms sequence and the B6 reference sequence.
2. In comparison with the B6 reference sequence, the MSM and JF1 sequences contain 38,182 and 38,124 non-synonymous SNPs in 11,489 and 11,313 genes, respectively.
3. Genome introgression from M. m. molossinus into M. m. domesticus is primary framework for the mosaic genomes of classical inbred strains.
4. The genomes of B6 and other classical inbred strains have long consecutive segments with extremely high similarity (>99.998%) to the JF1/Ms strain. This indicates that the ancestor of JF1/Ms was direct origin of M. m. molossinus genome in classical inbred strains.
5. Roughly 30 to 40% of the SNPs detected in pairwise comparisons of classical inbred strains are attributable to the M. m. molossinus genome introgression.
The sequence data of MSM/Ms and JF1/Ms are available through the NIG mouse genome database, with side-by-side comparison to the B6 reference sequence.
This work was carried out in collaboration with National Institute of Genetics (Mammalian Genetics Laboratory, Comparative Genomics Laboratory and Genome Biology Laboratory), RIKEN (BioResource Center and Genome Science Center) and the Wellcome Trust Sanger Institute, and was supported by a Grant-in-Aid for Scientific Research on Priority Areas “Comparative Genomics” and the National BioResource Projects “the Genome Information Upgrading Program” from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also supported in part by the Biodiversity Research Project of the Transdisciplinary Research Integration Center, Research Organization of Information and Systems.
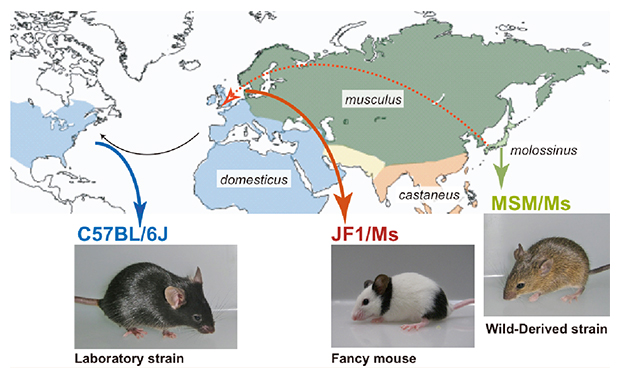
Three mouse strains mainly used in this study. C57BL/6J (B6) is a representative classical inbred strain, which was derived mostly from M. m. domesticus (west European subspecies). MSM/Ms and JF1/Ms originated from M. m. molossinus (Japanese subspecies).
A mouse model of human chromosomal disorder, partial trisomy distal 4q
Mammalian Genetics Laboratory・Shiroishi Group
Tamura, M., Hosoya, M., Fujita, M., Iida, T., Amano, T., Maeno, A., Kataoka, T., Otsuka, T., Tanaka, S, Tomizawa, S., and Shiroishi, T.
Human Molecular Genetics, 2013 Jun 15;22(12):2471-2481. DOI: 10.1093/hmg/ddt099
Human chromosomal disorders, such as trisomy or monosomy, occur in over 1% of newborns, and are often associated with developmental failures including congenital heart disease and mental retardation. Though these disorders are likely caused by imbalance in genes involving the trisomic or monosomic region, the molecular bases are unknown in most cases.
In this study, we demonstrated that Recombination-induced mutation 4 (Rim4) is a model animal of human chromosomal disorder, Partial trisomy distal 4q (4q+). Rim4 genome has extra fragment of 6.5Mb from mouse chromosome 8, which is syntenic to the distal end of human Chr4, 4q32.3 to 4q34.1, and contains 17 genes including basic helix-loop-helix transcription (bHLH) factor, Hand2. Rebalancing the gene dosage by genetic cross with Hand2 knockout mouse rescued symptoms of the heart and limb deformities of Rim4. These results suggest that over-dosage of Hand2 causes heart and limb deformities in Rim4 and 4q+, and Rim4 provides a unique animal model to understand the molecular bases underlying the complex phenotypes of 4q+.
This work was carried out in collaboration with National Institute of Genetics (Dr. Shiroishi group) and National Hospital Organization, Niigata Hospital (Dr. Tomizawa group).
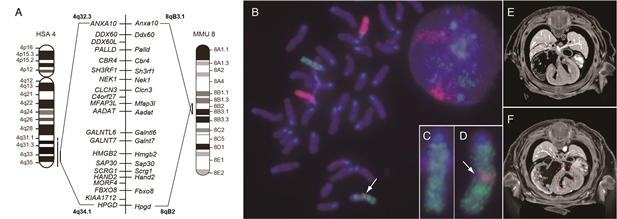
Chromosome aberration and ventricular septal defect observed in Rim4 mutant mouse.
(A) Mouse Chr8 (MMU) 8qB2–B3.1 is syntenic to human Chr4 (HSA4) 4q32.3–34.1, and genes and gene order are well conserved in the syntenic regions of the two species. (B) Dual-color whole chromosome FISH image of Chr6 (green color) and Chr8 (magenta color). Magnified images of Wild type and Rim4/+ Chr6 are shown in insets, (C) and (D), respectively. Arrow in (B) and (D) indicates Chr8-derived insertion fragment. μ-CT images of E14.5 embryos of wild type (E) and Rim4/Rim4 mouse (F) are shown. Dotted circle in (F) indicates ventricular septal defect.
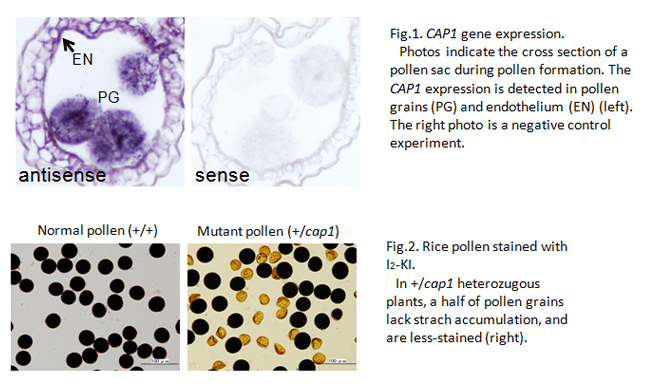
Rice gene essential for pollen formation
Experimental Farm・Nonomura Group
Kenji Ueda, Fumiaki Yoshimura, Akio Miyao, Hirohiko Hirochika, Ken-Ichi Nonomura, and Hiroetsu Wabiko
Plant Physiology, 2013 Jun;162(2):858-871. DOI: 10.1104/pp.113.216523
The genome of cultivated rice contains 32,000 genes, and more than 20,000 are expressed in developing anther and pollen. However, there are few genes whose function is evidenced in pollen development. In this study, we succeeded to identify the rice gene with its function specifically in pollen formation, and named COLLAPSED ABNORMAL POLLEN1 (CAP1).
In angiosperms, the pollen has a special, tricellular structure of a pair of sperm cells involved within the vegetative cell. The CAP1 gene is strongly expressed in anthers at the bicellular pollen stage (Fig.1). The pollen grains lacking CAP1 function lose most cellular components except for outer pollen wall, exine (Fig.2), and are unable to elongate the pollen tube at all. Homozygous cap1 mutant plants exhibit no remarkable aberration in other developmental stages, indicating the specific function of CAP1 in pollen development.
The amino acid alignment of rice CAP1 protein is similar to that of the plant L-arabinokinase. The function of this enzyme is supposed in phosphorylation of free L-arabinoses, generated during the cell-wall metabolism, for reuse in de novo wall formation. It is likely in rice cap1 mutant that the arabinoses unable to be reused accumulate aberrantly or the cell-wall metabolism is disrupted in pollen grains. One of Arabidopsis L-arabinokinase-like genes shows a similar expression pattern to rice CAP1-gene expression. This result suggests that the CAP1 function is conserved broadly among angiosperm species, and plays an important role in pollen development.
This is a collaborative work with the Akita Prefectural University and the National Institute of Agribiological Sciences, Japan, and is supported by the NIG Collaborative Research Funding.
Transcription factors interfering with dedifferentiation induce cell type-specific transcriptional profiles
Division of Molecular and Developmental Biology・Kawakami Group
Takafusa Hikichi, Ryo Matoba, Takashi Ikeda, Akira Watanabe, Takuya Yamamoto, Satoko Yoshitake, Miwa Tamura-Nakano, Takayuki Kimura, Masayoshi Kamon, Mari Shimura, Koichi Kawakami, Akihiko Okuda, Hitoshi Okochi, Takafumi Inoue, Atsushi Suzuki, and Shinji Masui
PNAS, 2013 Apr 16;110(16):6412-6417. DOI: 10.1073/pnas.1220200110
Transcription factors (TFs) are able to regulate differentiation related processes, including dedifferentiation and direct conversion, through the regulation of cell type-specific transcriptional profiles. However, the functional interactions between the TFs regulating different transcriptional profiles are not well understood. Here, we show that the TFs capable of inducing cell type-specific transcriptional profiles prevent the dedifferentiation induced by TFs for pluripotency. Of the large number of TFs expressed in a neural lineage cell line, we identified a subset of TFs that, when overexpressed, strongly interfered with the dedifferentiation triggered by the procedure to generate induced pluripotent stem cells. This interference occurred through a maintenance mechanism of the cell type-specific transcriptional profile. Strikingly, the maintenance activity of the interfering TF set was strong enough to induce the cell line-specific transcriptional profile when overexpressed in a heterologous cell type. Our results suggest that dedifferentiation suppresses a cell type-specific transcriptional profile, which is primarily maintained by a small subset of TFs capable of inducing direct conversion. We anticipate that this functional correlation might be applicable in various cell types and might facilitate the identification of TFs with induction activity in efforts to understand differentiation.
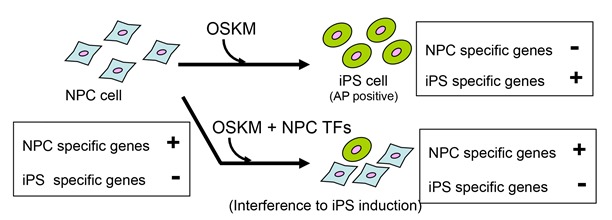
NPC (neural progenitor) cells are dedifferentiated by introduction of OSKM. This dedifferentiation was inhibited by expression n of NPC-specific transcription factors.
Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
Division of Cytogenetics・Kobayashi Group
Satoru Ide, Kimiko Saka, and Takehiko Kobayashi
PLOS Genetics, 2013 Apr;9(4):e1003410. DOI: 10.1371/journal.pgen.1003410
Gene amplification is one of the major strategies used by cells to increase the abundance of gene products. We have been studying amplification of the rDNA cluster in yeast with a focus on distinguishing the contributions of unequal sister-chromatid recombination vs rolling circle-type amplification as observed in the early developmental stage in amphibian oogenesis. It is not known how these two modes of amplification switch. We screened for yeast mutants in which rDNA copy number was unstable and found and characterized an rtt109 mutant that has an abnormally high rDNA copy number. Evidence is presented that the rolling circle-type amplification occurs in this mutant. Therefore, RTT109 plays a key role in regulating mode of rDNA amplification.
Variation in gene copy number (amplification) has been widely detected in various organisms, contributing to both beneficial adaptation and pathology (e.g., cancer). Our results shed new light on molecular mechanisms of gene amplification.
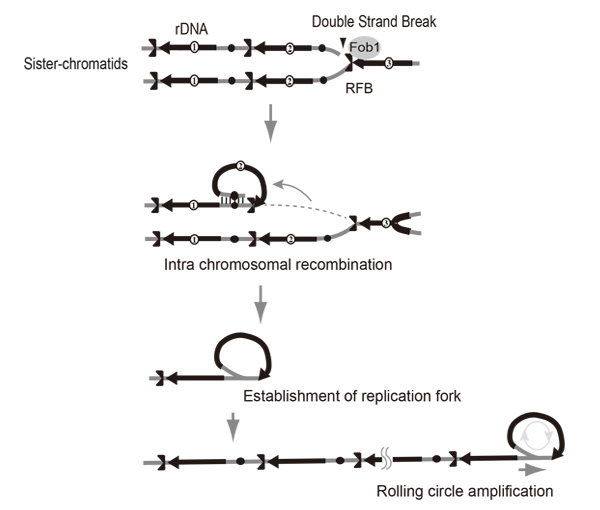
Rolling circle replication by intra-sister chromatid recombination.The broken end at the RFB (Replication Fork Barrier site) recombines via intra-sister chromatid exchange followed by rolling circle replication.
Neocentromere formation using chromosome engineering
Press release
Chromosome engineering allows the efficient isolation of vertebrate neocentromeres
Wei-Hao Shang, Tetsuya Hori, Nuno M.C. Martins, Atsushi Toyoda, Sadahiko Misu, Norikazu Monma, Ichiro Hiratani, Kazuhiro Maeshima, Kazuho Ikeo, Asao Fujiyama, Hiroshi Kimura, William C. Earnshaw, and Tatsuo FukagawaDevelopmental Cell, 24(6), 635-648, 14 March 2013. DOI: doi:10.1016/j.devcel.2013.02.009
Centromeres are specified by sequence-independent epigenetic mechanisms in most organisms. Rarely, centromere repositioning results in neocentromere formation at ectopic sites. However, the mechanisms governing how and where neocentromeres form are unknown.
Here, we established a chromosome-engineering system in chicken DT40 cells that allowed us to efficiently isolate neocentromere-containing chromosomes.
Neocentromeres appear to be structurally and functionally equivalent to native centromeres. Chromatin immunoprecipitation sequencing (ChIP-seq) analysis with 18 neocentromeres revealed that the centromere-specific histone H3 variant CENP-A occupies an ∼40 kb region at each neocentromere, which has no preference for specific DNA sequence motifs. Furthermore, we found that neocentromeres were not associated with histone modifications H3K9me3, H3K4me2, and H3K36me3 or with early replication timing. Importantly, low but significant levels of CENP-A are detected around endogenous centromeres, which are capable of seeding neocentromere assembly if the centromere core is removed.
Our experimental system provides valuable insights for understanding how neocentromeres form. This was performed by Wei-Hao Shang and Tetsuya Hori (Fukagawa Lab) in collaboration with TRIC of ROIS, Fujimaya Lab. (NIG, NII), Maeshima Lab (NIG), Ikeo Lab. (NIG), Kimura Lab (Osaka U.), and Eranshaw Lab (U. Edinburgh).
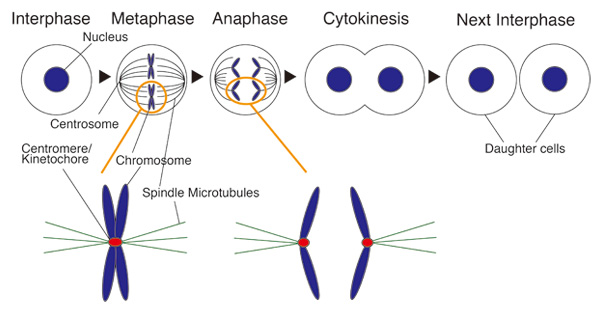
Cell division and centromere
During mitosis spindle microtubules capture a special structure of chromosome for faithful chromosome segregation. This structure is called “Kinetochore”. Centromere is a genome region in which kinetochore is formed. Dysfunction of kinetochore results in some diseases including cancer.
Innervation is required for sense organ development
Division of Molecular and Developmental Biology・Kawakami Group
Hironori Wada, Christine Dambly-Chaudiere, Koichi Kawakami, and Alain Ghysen
PNAS, 2013 Apr 2;110(14):5659-5664. DOI: 10.1073/pnas.1214004110
Various types of sense organs are distributed over the body surface. These organs are innervated by sensory axons, thereby transmit information to the central nervous system. Many sense organs emit molecules required for proper growth or guidance of the axons. However, roles of the axons for development of sense organs remain poorly understood. In this study, we reveal that proliferation of the mechanosensory organs (neuromasts) of fish is promoted by axonal innervation.
In adult zebrafish, the neuromasts give rise to new neuromasts by budding and generate a cluster of organs. The budding cells, that show high Wnt signaling activity, are associated by side branches of axons extended from the founder neuromast (Fig. 1A). To analyze the role of innervation, we ablated sensory neurons in larvae. The neuromast sent off budding cells normally, but subsequent cell proliferation to generate new sense organs did not take place (Fig. 1B,C). We propose that the axon promotes Wnt signaling activity, which is required for the proliferate phase that leads to sense organ formation.
This study was carried out in collaboration with Dr. Alain Ghysen (Montpellier University) and funded by the PRESTO of the Japan Science and Technology Agency.
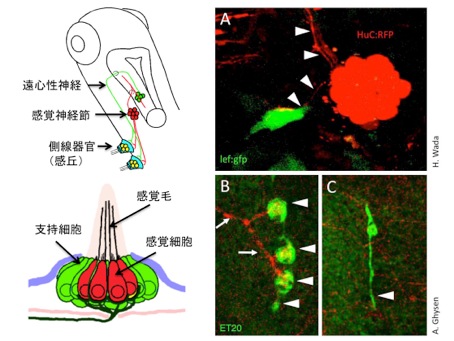
(A) The lateral line sense organ, neuromast, sends off proliferative bud cells, that show high Wnt signaling activity. Axons are indicated by arrowheads.
(B, C) The cell proliferation does not take place in the absence of the sensory axons.
Seeing the visual world in the brain in real time
Press release
Real-Time Visualization of Neuronal Activity during Perception
Akira Muto, Masamichi Ohkura, Gembu Abe, Junichi Nakai and Koichi Kawakami Current Biology, 23(4), Feb 18, 2013. DOI: 10.1016/j.cub.2012.12.040The visual world is first projected onto the retina, and the visual information is further transmitted to the brain. In this initial stage of visual processing, the visual world is mapped on the brain, which is called visuotopy. This is a common feature found in the brains of all animals with eyes. While visuotopy is a well-established notion, no one has ever demonstrated this in real time in a natural condition. In this study we developed an improved version of GCaMP, a calcium sensor, in collaboration with Prof. Nakai at Saitama University. Using this new highly sensitive calcium probe, we could visualize neuronal activity in a zebrafish larva during visual perception in prey capture behavior.
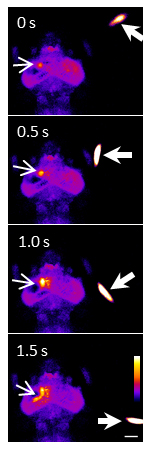
A swimming paramecium (arrowheads) evoked Ca2+ transients (arrows) in the neuropil and cell bodies of the left tectum of a one-week old larva embedded in agarose. Ratio images were created and pseudo-colored. Scale bar represents 100 μm.
▶This method is one of the basis of these researches. A virtual reality system to analyze neural activity and behavior in adult zebrafish Appetite control via hunger and satiety Neural signatures of sleep in zebrafish Glia-neuron interactions underlie state transitions to generalized seizures ‘Eating with the eyes’ is hard-wired in the brain
Morphogenesis of bacterial cells analyzed by a high performance sequencer
Microbial Genetics Laboratory・Niki Group
Comparative Genomics Laboratory・Fujiyama Group
Genome Biology Laboratory・Kohara Group
Wild-type E. coli are rod shape (Figure A, left). To make rod shaped cells, it is necessary that many proteins form complexes and function properly. E. coli cells are surrounded by rigid peptidoglycan (PG) layer and have to synthesize PG correctly. So far, we have identified RodZ required for determination of cell shape.
RodZ-deficient mutant is round shape (Figure A, middle) and grows slower than wild-type E. coli cell. To reveal function of RodZ protein, we isolated suppressor mutants that restore cell growth and shape of the rodZ mutant. To map the mutation sites in the suppressor mutants, we sequenced whole genome of twenty-nine mutants by a next-generation sequencer Solexa. This is the first report that mutation sites in ~30 mutants are determined by whole genome sequencing.
Most of the mutations were found in mreB, mrdA, or mrdB genes. It has been hypothesized that MreB, PBP2 encoded by mrdA gene, and RodA encoded by mrdB gene function with RodZ. Especially, twenty of twenty-nine mutants had a mutation in mreB gene. In addition, these mutations were clustered in domain 1A, one of the subdomain of MreB protein. These mutations change properties of MreB protein so that E. coli can form rod shape without RodZ protein. We also found that mutants of PBP2 and RodA change properties of MreB. Thus, we concluded that RodZ regulates function of MreB to form rod shape of E. coli.
This work was done by a collaboration of Niki lab, Fujiyama lab, and Kohara lab.
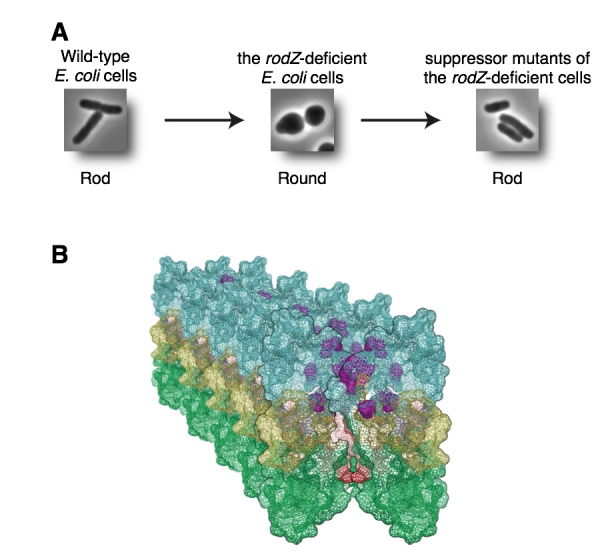
(A) Wild-type E. coli is rod (left). RodZ-deficient E. coli cell is round (middle). Suppressor mutants are rod (right). (B) Mutation sites shown by purple are clustered in domain 1A and it is a surface between MreB filaments. These mutations change properties of MreB filaments.
Structural core for kinetochore formation
Division of Molecular Genetics・Fukagawa Group
The kinetochore forms a dynamic interface with microtubules from the mitotic spindle during mitosis. The Ndc80 complex acts as the key microtubule-binding complex at kinetochores. However, it is unclear how the Ndc80 complex associates with the inner kinetochore proteins that assemble upon centromeric chromatin. Here, based on a high-resolution structural analysis, we demonstrate that the N-terminal region of vertebrate CENP-T interacts with the “RWD” domain in the Spc24/25 portion of the Ndc80 complex. Phosphorylation of CENP-T strengthens a cryptic hydrophobic interaction between CENP-T and Spc25 resulting in a phospho-regulated interaction that occurs without direct recognition of the phosphorylated residue. The Ndc80 complex interacts with both CENP-T and the Mis12 complex, but we find that these interactions are mutually exclusive, supporting a model in which two distinct pathways target the Ndc80 complex to kinetochores. Our results provide a model for how the multiple protein complexes at kinetochores associate in a phospho-regulated manner.
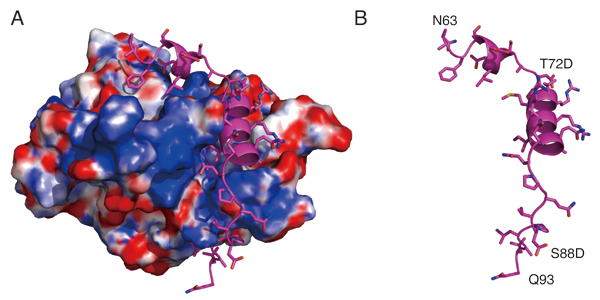
Structural model showing the surface charge of the Spc24/25 complex interacting with phospho-mimetic CENP-T peptide (Cyan). (B) Structural model showing the phospho-mimetic CENP-T peptide from the CENP-T-Spc24/25 complex structure in (A) on its own.
Takehiko Kobayashi, Professor of Cytogenetics, was awarded “the 29th Inoue prize” by Inoue Foundation for Science.

 Title
Study on gene amplification and its relationship to cancer and aging
Takehiko Kobayashi, Professor of Cytogenetics, was awarded “the 29th Inoue prize” by Inoue Foundation for Science. This prize is awarded to scientists who conducted basic and original scientific works. Takehiko Kobayashi revealed the molecular mechanism of “gene amplification” that contributed to cancer and aging.
Inoue Foundation for Science
※This site is presented only in Japanese.
Title
Study on gene amplification and its relationship to cancer and aging
Takehiko Kobayashi, Professor of Cytogenetics, was awarded “the 29th Inoue prize” by Inoue Foundation for Science. This prize is awarded to scientists who conducted basic and original scientific works. Takehiko Kobayashi revealed the molecular mechanism of “gene amplification” that contributed to cancer and aging.
Inoue Foundation for Science
※This site is presented only in Japanese.Identification of male promoting signal in mouse germ cells
Mammalian Development Laboratory・Saga Group
In the mouse, testicular development is triggered in somatic cells by the function of Sry followed by the activation of fibroblast growth factor (FGF) 9, which regulates testicular differentiation in both somatic and germ cells. However, the mechanism is unknown. We show here that Nodal/Activin signaling pathway is activated in both male gem cells and somatic cells. The disruption of Nodal/Activin signaling drives male germ cells into meiosis and causes ectopic initiation of female-specific genes in somatic cells. Furthermore, we prove that Nodal/Activin-A works directly on male germ cells to induce male specific gene, Nanos2 independently of FGF9. We conclude that Nodal/Activin signaling is required for testicular development and propose a model in which Nodal/Activin-A acts downstream of FGF signaling to promote male germ cell fate and protect somatic cells from initiating female differentiation.
This study is conducted by Quan Wu who is a current student of SOKENDAI.
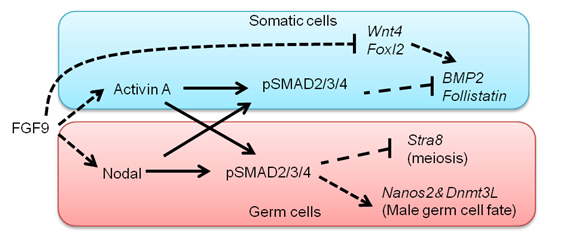
A model proposed by our study. FGF signals activate Nodal/Activin signaling pathway in both somatic cells and germ cells. Nodal/Activin-A triggers male sex differentiation by inducing male-specific genes, Nanos2 and Dnmt3L. Meanwhile, it suppresses Stra8 that is an essential gatekeeper of meiosis. In somatic cells Nodal/Activin-A thwarts the process of female differentiation by inhibiting Bmp2 and Follistatin.

















