Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Katsuhiko Minami wins the “Early Career Award in Biophysics” and “IUPAB Award” at the 63rd Annual Meeting of the Biophysical Society of Japan

Katsuhiko Minami, a postdoctoral researcher in the Genome Dynamics Laboratory (Maeshima Lab), received the 21st Early Career Award in Biophysics at the 63rd Annual Meeting of the Biophysical Society of Japan, held in Nara, Japan, on September 24–26, 2025. Additionally, he was presented with the IUPAB Award from the International Union of Pure and Applied Biophysics (IUPAB), given to the most outstanding awardee.
▶ Awarded presentation title: Replication-dependent histone (Repli-Histo) labeling dissects the physical properties of euchromatin/heterochromatin in living human cells
▶ Early Career Award in Biophysics | The Biophysical Society of Japan
▶ The 63rd Annual Meeting of the Biophysical Society of Japan
Convergent losses of arbuscular mycorrhizal symbiosis in carnivorous plants
Press release
Convergent losses of arbuscular mycorrhizal symbiosis in carnivorous plants
Héctor Montero, Matthias Freund, Kenji Fukushima
New Phytologist (2025) DOI:10.1111/nph.70544
Most land plants form the ancient arbuscular mycorrhizal (AM) symbiosis, while carnivory is a younger trait that evolved in several angiosperm orders. The two biotic interactions similarly help plants acquire mineral nutrients, raising the question of whether they can coexist. However, the mycorrhizal status of carnivorous plants has long remained speculative. We surveyed the occurrence of AM-associated genes across carnivorous plant lineages, performed AM fungal inoculation assays, and microscopically evaluated the patterns of colonization. We found convergent losses of the AM trait either coincident with or predating the emergence of carnivory. Exceptionally, the carnivorous plant Roridula gorgonias retains symbiosis-related genes and forms arbuscules. The youngest carnivorous lineage, Brocchinia reducta, showed signatures of the early stages of AM trait loss. An AM-associated CHITINASE gene encodes a digestive enzyme in the carnivorous plant Cephalotus, suggesting gene cooption. We uncovered a mutually exclusive trend of AM symbiosis and carnivory, with only rare instances of coexistence. These findings illuminate the largely unexplored processes by which plant nutritional strategies evolve and supplant one another over time.

Photo: Brocchinia reducta growing on Mt. Roraima in the Guiana Highlands (photo by Andreas Fleischmann)
Guideline for Application for 2026 NIG-JOINT(Joint Research and Research Meeting) (*Application was closed)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Islam, Moutushi, D5 SOKENDAI student received the Morishima Award

Ms. Islam, Moutushi (Molecular Cell Engineering Laboratory, Kanemaki Laboratory) received the Morishima Award from the Genetics Program at SOKENDAI. At the award ceremony held on September 19, 2025, Dr. Saito, Chair of the Genetics Program, presented them with certificates and a catalog of the research grant.
The Morishima Award is given to students in the Genetics Program to honor their outstanding performance during their PhD studies and to encourage further achievements.
・Islam, Moutushi (Molecular Cell Engineering Laboratory, Thesis Advisor: Kanemaki, Masato)
・thesis title : Advancing the utility of AID2-based conditional protein knockdown
Under what conditions does speciation occur?
The genomics of discrete polymorphisms maintained by disruptive selection
Jun Kitano*, Kotaro Kagawa, Takashi Tsuchimatsu, Ryo Yamaguchi, Masato Yamamichi *corresponding author
Trends in Ecology & Evolution (2025) 40 DOI:10.1016/j.tree.2025.08.003
Disruptive selection can lead to the evolution of discrete morphs. We show that particular genetic architectures, in terms of dominance, epistasis, and linkage, are likely to evolve to produce discrete morphs under disruptive selection. Recent genomic studies have revealed that causative mutations tend to cluster, sometimes as a result of chromosomal rearrangements, but we still know little about the molecular mechanisms of dominance and epistasis. Although disruptive selection can also lead to speciation, once an optimal genetic architecture has evolved, disruptive selection no longer promotes the evolution of assortative mating. For a better understanding of the conditions that promote or constrain speciation, it is necessary to address how fast such a genetic architecture can evolve.
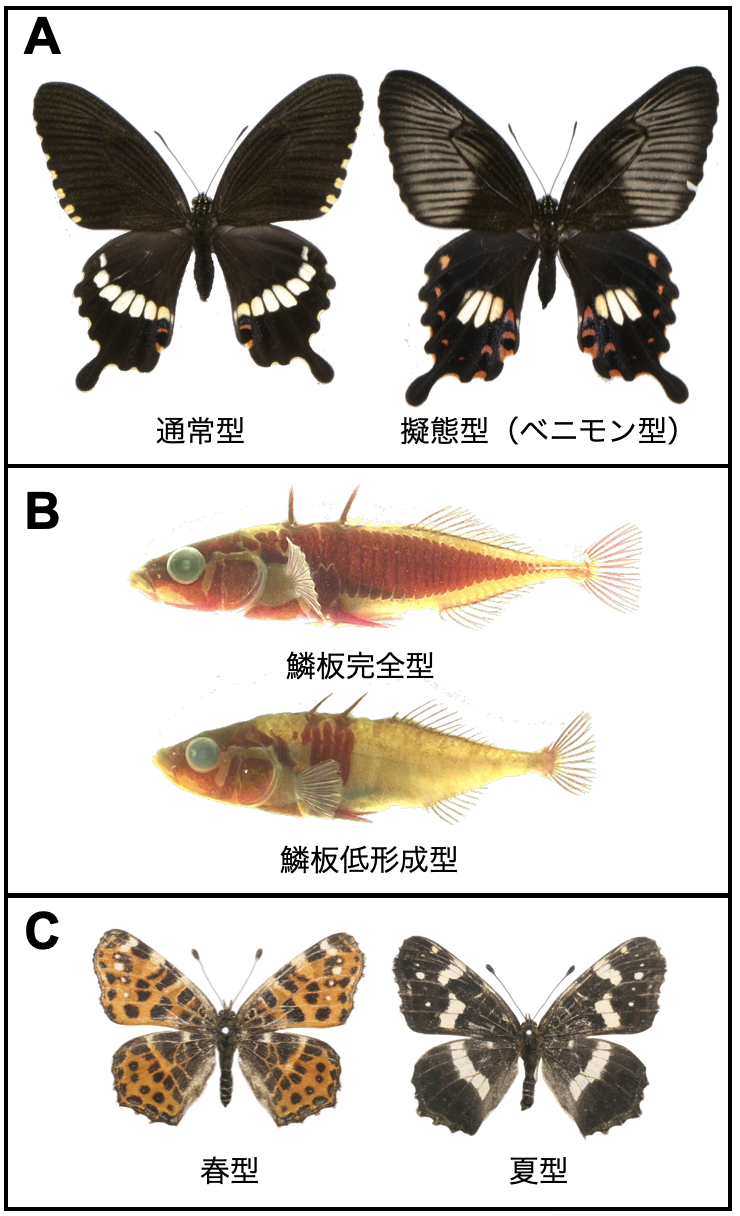
Figure: Although they look different, they are all polymorphic forms within the same species.
Postdoctoral PositionLaboratory of Gene Quantity Biology (PI: Mariko Sasaki)
Our laboratory investigates the fundamental mechanisms of genome instability, gene amplification, and circular DNA in cancer. Gene amplification is a major driver of tumorigenesis and occurs as a consequence of genome instability. It was traditionally believed to occur on chromosomes. However, it is now clear that gene amplification also involves the formation and accumulation of extrachromosomal DNA (ecDNA) —circular DNA derived from chromosomal fragments carrying oncogenes or drug-target genes. ecDNA has been found in approximately 17% of cancer patient samples and is increasingly recognized as a key factor promoting cancer development, progression, and therapy resistance. Importantly, despite its chromosomal origin, ecDNA acquires unique properties, including distinct chromatin structures, gene expression patterns, and unconventional segregation patterns. Nevertheless, many fundamental aspects of ecDNA biology remain poorly understood. Our laboratory aim to answer central questions such as:
- What molecular mechanisms drive the formation of ecDNA?
- What new properties does ecDNA acquire through circularization?
- (with particular focus on DNA replication, recombination, and repair)
- How does ecDNA evolve in cancer cells to promote tumor progression and therapy resistance?
By addressing these questions and by exploring applications that leverage ecDNA-driven gene amplification, we pursues both basic and translational research toward innovative cancer therapies. We also study other types of circular DNA present in eukaryotic cells to unravel the new biology driven by circular DNA. We are seeking postdoctoral researchers who have solid backgrounds in fields such as molecular biology, genetics, biochemistry, cell biology, biophysics, or genome biology.
National Institute of Genetics (NIG) is an internationally recognized research institute that attracts outstanding scientists worldwide. English is the primary language in seminars and discussions, world-leading scientists are frequently invited to give seminars, providing an excellent setting for developing a global network.
National Institute of Genetics (NIG) is an internationally recognized research institute that attracts outstanding scientists worldwide. English is the primary language in seminars and discussions, world-leading scientists are frequently invited to give seminars, providing an excellent setting for developing a global network.
We are looking for a talented, highly motivated, and enthusiastic posdoc. We welcome applicants who already have or are willing to obtain their own fellowships or research fundings in the future. Please also note that NIG holds an annual open call for its own postdoctoral fellowship program, although the application for the yeast of 2026 is now closed.
Contract information
Duration: 2 years with annual evaluation at the end of fiscal year
Working time: full time
Benefits: according to NIG regulations
Location: Mishima city, Shizuoka prefecture, Japan
Please send CV, summary of your current research project (1–3 pages), letter of motivation, and at least 2 references to . Applicants may be later asked to submit a research proposal (although the research project that the applicant will actually work on will be decided after discussions).
Call For Application for Project Associate Professor Position of NIGiRC (Integrated Research Core)
The National Institute of Genetics (herein “NIG”) invites applications for Project Associate Professor positions who are able to carry out independent research based on the creative ideas. The research cycle of life science consists of several steps, including findings based on observation and measurement data, making of a new hypothesis, and empirical tests of the hypothesis. The NIGiRC (Integrated Research Core) is established for the purpose of accelerating and enhancing this research cycle, and NIG seeks for Project Associate Professors who can proceed with innovative research and contribute toward the goal of NIGiRC.
Application Guidelines:PDF Application Materials:Brief summary of career(excel)
1. Recruiting: Project Associate Professor (2 qualified persons)
2. Appointment Start Date: As soon as possible after the selection decision is made
3. Term of Appointment: 5 years
4. Qualifications:
(1) Young researchers who obtained Ph.D. within the last 13 years as of April 1, 2025 (For those who suspended their research activities due to life events, the suspension period is not included in the calculation of the total years after obtaining Ph.D.) and who are able to carry out research independently based on their own ideas. Any research field is acceptable.
(2) Those who can get basic funding for conducting their own research
(3) Each applicant must appoint one NIG Professor who has five or more years left until his/her retirement as of April 1. 2025 as a mentor (Please obtain his or her prior agreement to act as mentors).
5. Background for Recruitment:
NIGiRC (Integrated Research Core) is newly established by integrating NIG’s three independently operated centers (“Genetic Resource Center”, “Phenotype Research Center” and “Advanced Genomics Center”). NIGiRC is going to be operated in collaboration with “BSI: BioData Science Initiative” which was installed within the Joint Support-Center for Data Science Research (ROIS-DS) in FY2022. The aim of establishment of NIGiRC is to accelerate the research cycle in life science across various fields in Japan. At the time of its establishment, NIG employs qualified researchers.
6. Responsibilities:
Each appointed researcher is to carry out independent research under the mentoring of NIG Professor.
7. Research environment and support:
Each appointed researcher will use the space and facilities of the laboratory of the mentor professor.
He or she will be additionally provided with an individual research room of approx. 19 ㎡. In addition to that, as an Inter-University Research Institute, NIG provides substantial common equipment and research infrastructure, and so appointed researchers can make best use of NIG’s resource on hand to conduct their research activities. Therefore, it is not necessary to prepare all research equipment on their own from the start. Appointed researchers will receive research funds of about 1 million yen per year.
8. Wages: Based on ROIS regulations
9. Social Insurances: MEXT Mutual Society of Health Insurance / Worker’s Accident Compensation Insurance / Employment Insurance
10. Work hours: Discretionary Labor System (Deemed working hours: 7 hours 45 minutes/day)
11. Holiday: Saturday and Sunday, National Holidays, From December 29th to January 3rd (New Year’s Break), Summer Break
12. Application Deadline: Noon (Japan Time) on October 10, 2025
13. Application Materials:
I. Curriculum Vitae (Write in English or Japanese / Free form / Write years in Western calendar)
II. List of publications and grants (Write in English or Japanese / Free form / If there are multiple authors, briefly describe your contribution / Indicate a few most significant research papers / Regarding the research grants, describe whether you are PI or co-PI)
III. Summary of past and present research and future directions (Write in English or Japanese / Free form / Length is about three A4 pages / Include figures if necessary.)
IV. Names and contacts of three or more references and the name of the NIG Professor whom the applicant appoints as a mentor
V. Summary of career
VI. Your key papers (in PDF file / up to three papers)
*All the personal data in the application is handled in strict confidentiality, and used solely for recruitment purposes.
14. Submission:
Submit items I to VI listed above electronically as follows:
(a) The subject line should be “Application for Project Associate Professor positions in the NIGiRC”, which should be also noted in the e-mail body.
(b) Put items I to IV together in a single file by separating each item by page. E-mail the file as an attachment. The file format should be MS Word or PDF. Also attach item V (form can be downloaded from the NIG homepage) as a separate file.
(c) Send your key papers as PDF attachments. The file format should be PDF. If your key papers are available to access online, include a list of URLs in the e-mail body.
* We will notify you via e-mail upon receipt of your electronic application within two business days.
Based on gender equality, National Institute of Genetics has been actively promoting female scientists. In case of equivalent aptitude and achievements in research, education, and social contributions, preference will be given to female candidates.
The National Institute of Genetics is promoting safety and healthy working, including preventing passive smoking. (Indoor smoking is banned. / Smoking is allowed only in the designated outdoor areas.)
Send your application and inquiries to:
NIG Personnel Committee (Personnel Team) E-mail:
Mailing Address: National Institute of Genetics
Yata 1111, Mishima, Shizuoka 411-8540 JAPAN
TEL: +81-55-981-6716 FAX: +81-55-981-6715
Homepage: https://www.nig.ac.jp/nig/
NIG Organization Chart: https://www.nig.ac.jp/nig/research/organization-top/organization
15. Selections:
(1) Preliminary Screening (Document Review)
Applicants will be shortlisted based on their submitted documents.
The selection result will be e-mailed directly to applicants by the beginning of November 2025.
Please contact the Personnel Team (Refer to 14. Submission) if you do not receive e-mail by that time.
(2) Secondary Screening (Interview)
Shortlisted candidates will be invited to an interview. They are required to give a short presentation about their past research and future plan at our institute. Subsequently, a question and answer session will be carried out. Interviews will be carried out in November or December 2025. In principle, applicants in Japan are requested to attend an interview at NIG (Travel expenses will be provided). For those living overseas, interviews will be basically conducted online via Zoom, etc.
New assistant professor joins NIG
New assistant professor joins NIG as of September 1, 2025.
▶ NAKAJO, Haruna : Asakawa Group • Neurobiology and Pathology Laboratory
Bharathi Venkatachalam won the “Neuroscience2025 Excellence Award for the Training School for Next Generation Scientists”
Bharathi Venkatachalam, D4 student in Mouse Genomics Resource Laboratory(Koide Laboratory), received the “Neuroscience2025 Excellence Award for the Training School for Next Generation Scientists” at the “The 48th Annual Meeting of the Japanese Neuroscience Society” held in Niigata, Japan, on July 24th – 27th.
▶ Awarded presentation title: Neural mechanism underlying motivation to approach humans in mice showing active tameness
▶ The 48th Annual Meeting of the Japanese Neuroscience Society

Bharathi Venkatachalam















