Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
A cell suicide module by use of the auxin-inducible degron (AID) technology
Molecular Function Laboratory • Kanemaki Group
A cell suicide module: Auxin-induced rapid degradation of Inhibitor of Caspase Activated DNase (ICAD) induces apoptotic DNA fragmentation, caspase activation and cell death
Kumiko Samejima, Hiromi Ogawa, Alexander V. Ageichik, Kevin L. Peterson, Scott H. Kaufmann, Masato T. Kanemaki and William C. Earnshaw The Journal of Biological Chemistry, 298, 31617-31623, 2014.; DOI:10.1074/jbc.M114.583542Caspase-activated DNase (CAD), a DNase responsible for DNA fragmentation in apoptosis, is activated upon degradation of its inhibitor ICAD. However, it has not been known whether ICAD degradation is sufficient for induction of apoptosis. To answer this question, we established DT40 cells, in which ICAD can be degraded by addition of auxin using the auxin-inducible degron system. Upon rapid degradation of ICAD, we showed that DT40 cells were induced for apoptosis, indicating that ICDA degradation is sufficient for apoptosis. Moreover, this cell-killing system works in budding yeast cells. We suggest that the cell-killing system we established in this work might be applicable for containment measures of gene-modified organisms in the future.
This study was carried out as collaboration with Dr. Kumiko Samajima and Professor William Earnshaw in University of Edinburgh, UK. Dr. Earnshaw is also an invited professor of NIG.
た。
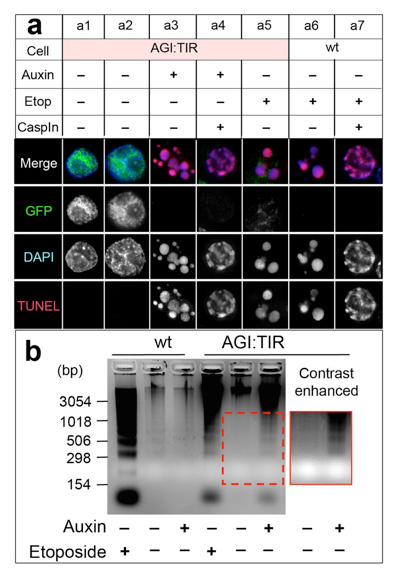
By adding auxin, ICAD was rapidly degraded, resulting induction of apoptosis (a3). In these cells, genomic DNA was fragmented (b).
Naruya SAITOU, Professor of Division of Population Genetics, was awarded an honorary doctorate from Université Toulouse III
Established in 1220, Université Toulouse III is one of the oldest and prestigious Universities. The university is also known as Paul Sabatier University after Paul Sabatier who received a Nobel Prize in Chemistry with François Auguste Victor Grignard in 1912.
Three other recipients are from Belgium, the United States, and Spain.
Division of Population Genetics • Saitou Group

Professor SAITOU, Naruya

Docteur Honoris Causa
Pradeep Lal (Postdoctoral Researcher at Division of Molecular and Developmental Biology Laboratory) received Poster Award

right:Pradeep Lal

BestPosetrAward
Division of Molecular and Developmental Biology • Kawakami Group
A simple method for generation of yeast conditional mutants by use of the auxin-inducible degron technology
Molecular Function Laboratory • Kanemaki Group
Rapid Depletion of Budding Yeast Proteins via the Fusion of an Auxin-Inducible Degron (AID)
Kohei Nishimura and Masato T. Kanemaki Current Protocols in Cell Biology, 64, 20.9.1-20.9.16, 2014; DOI:10.1002/0471143030.cb2009s64Budding yeast has been an important eukaryotic model organism because there have been many genetic technologies available. In the yeast research field, temperature-sensitive (ts) mutants have been generated in order to inactivate a target protein conditionally. Although ts-mutants have been useful, there are problems as well; a drastic temperature shift causes heat-shock responses and there are some cases in which a target protein is not inactivated in a short period of time. To overcome these problems, we transplanted a plant-specific degradation pathway controlled by the plant hormone auxin to budding yeast and, in such yeast cells, a protein of interest can be degraded upon addition of auxin. We named this technology as the auxin-inducible degron (AID) system (Nishimura et al, Nature Methods, 2009) and subsequently improved it by making efficient degron cassettes (Kubota et al. Molecular Cell, 2013). The AID technology is getting popular in the filed of yeast studies. In this paper, we described a detailed protocol for generation of budding yeast AID mutants by one transformation. All plasmids and yeast strains required for the experiment are available from NBRP (http://yeast.lab.nig.ac.jp/nig/index_en.html). We hope that this protocol would help your study in the future.
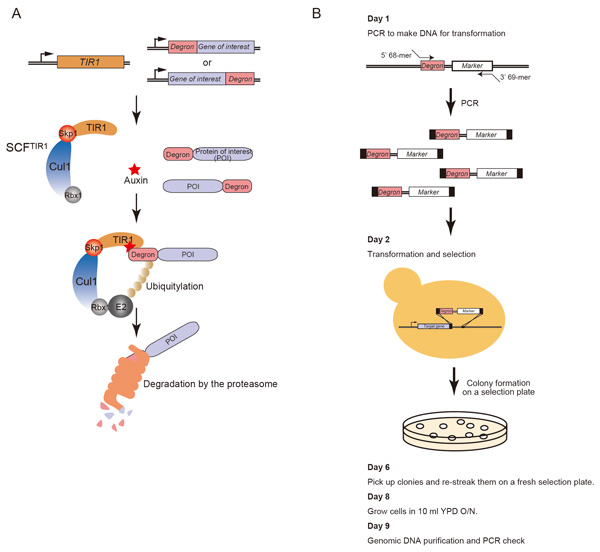
(A) Schematic illustration showing the mechanism of the auxin-dependent degradation of a degron-fused protein. (B) Schematic illustration showing the protocol for generation of AID mutants.
Chromatin foundation for the kinetochore machinery
Division of Molecular Genetics • Fukagawa Group
The centromere: chromatin foundation for the kinetochore machinery
Tatsuo Fukagawa, and William C. Earnshaw Dev. Cell 30,496-508,2014; DOI:10.1016/j.devcel.2014.08.016Since discovery of the centromere-specific histone H3 variant CENP-A, centromeres have come to be defined as chromatin structures that establish the assembly site for the complex kinetochore machinery. In most organisms, centromere activity is defined epigenetically, rather than by specific DNA sequences. In this review, Fukagawa (Dep. Mol. Genet.) and Earnshaw (ex visiting professor of NIG, Edinburgh Univ) describe selected classic work and recent progress in studies of centromeric chromatin with a focus on vertebrates. We consider possible roles for repetitive DNA sequences found at most centromeres, chromatin factors and modifications that assemble and activate CENP-A chromatin for kinetochore assembly, and use of artificial chromosomes and kinetochores to study centromere function.
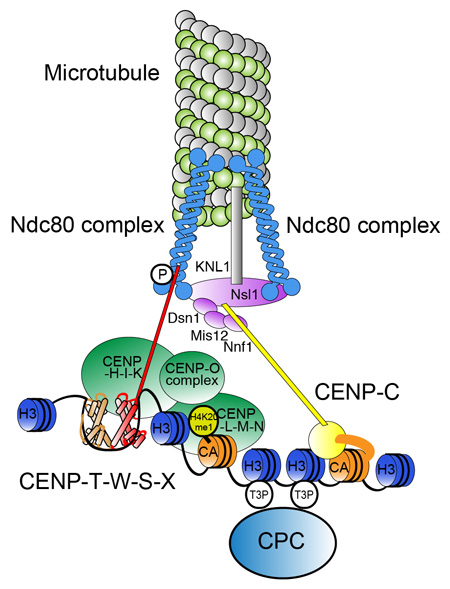
Molecular architecture of natural kinetochores. Centromere chromatin including is crucial for centromere specification and kinetochore assembly at natural centromeres. At the base of the structure are CENP-A containing nucleosomes, centromere specific H3 nucleosomes, and a CENP-T-W-S-X nucleosome-like structure in centromere chromatin. Centromere-specific chromatin structure is established by coordination of these components. CCAN proteins assemble on the centromeric chromatin and the microtubule-binding complex is subsequently recruited to assemble the functional kinetochore.
Structural study of the homology domain of the eukaryotic DNA replication proteins Sld3/Treslin
Biomolecular Structure Laboratory • Shirakihara Group Division of Microbial Genetics • Araki Group
Crystal structure of the homology domain of the eukaryotic DNA replication proteins Sld3/Treslin
Hiroshi Itou, Sachiko Muramatsu, Yasuo Shirakihara, and Hiroyuki Araki Structure Published: August 7, 2014 DOI:10.1016/j.str.2014.07.001The initiation of eukaryotic chromosomal DNA replication requires formation of an active replicative helicase at the replication origins of chromosomal DNA. Yeast Sld3 and its metazoan counterpart Treslin are the hub proteins mediating protein associations critical for the helicase formation. In this study we show the crystal structure of the central domains of Sld3 that is conserved in Sld3/Treslin family proteins. The domain consists of two segments with 12 helices and is sufficient to bind to Cdc45, the essential helicase component. The structural model of the Sld3-Cdc45 complex that is crucial for formation of the active helicase was proposed.
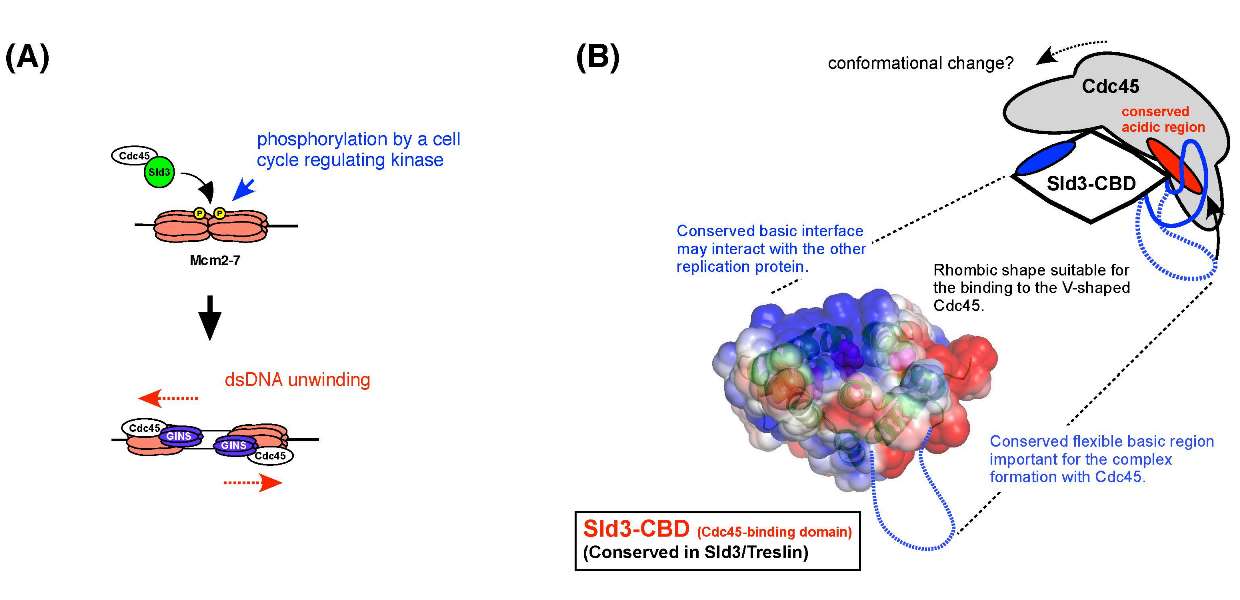
(A) Sld3 and Cdc45 form a complex that associates with origins in a mutually dependent manner,and its association to origins depends on the phosphorylation of the Mcm2-7 helicase core complex. Further protein interactions through Sld3 recruits GINS, another essential component of the active helicase. (B) Representation of the tertiary structure of the homology domain shared by Sld3 and Treslin. The proposed model of Sld3-Cdc45 complex is also shown.
Mr. Akihiro Uzuka (D2 Student SOKENDAI at Symbiosis and Cell Evolution Laboratory) received Poster Award.
 Mr. Akihiro Uzuka (D2 Student SOKENDAI at Symbiosis and Cell Evolution Laboratory) made a poster presentation at “Biology of plastids – towards a blueprint for synthethic organelles” which was held at Pultusk in Poland, 21-26 June 2014. And he received Poster Award for his poster presentation.
Mr. Akihiro Uzuka (D2 Student SOKENDAI at Symbiosis and Cell Evolution Laboratory) made a poster presentation at “Biology of plastids – towards a blueprint for synthethic organelles” which was held at Pultusk in Poland, 21-26 June 2014. And he received Poster Award for his poster presentation. Poster
PosterAnalyses of photosynthetic oxidative stress responses in herbivorous unicellular organisms
 HP
HPBiology of plastids -towards a blueprint for synthethic organelles
Axon elongation and guidance supported by M6 membrane proteins
Division of Brain Function • Hirata Group
Transcallosal Projections Require Glycoprotein M6-Dependent Neurite Growth and Guidance.
Sakura Mita, Patricia de Monasterio-Schrader, Ursula Fünfschilling, Takahiko Kawasaki, Hidenobu Mizuno, Takuji Iwasato, Klaus-Armin Nave, Hauke B. Werner, and Tatsumi Hirata. Cerebral Cortex DOI: 10.1093/cercor/bhu129Four-transmenbrane protein family M6 is concentrated on axon tips. Although in vitro studies have suggested its involvement in axon elongation, in vivo evidence has been lacking. In this study, we made double knockout mice for two family members M6a and M6b, and showed that these membrane proteins in fact control axon elongation in vivo. In the M6a/M6b double knockout brain, the corpus callosum, a long axon commissure that connects left-right brain hemispheres, was severely hypoplastic; many axons were found to stop short of reaching the midline. Furthermore, a fraction of callosal axons were misrouted subcortically. These results show that M6 proteins are indeed important regulators of axon growth and guidance in the developing brain.
Superenova system developed by the Division of Neurogenetics is used in this study.
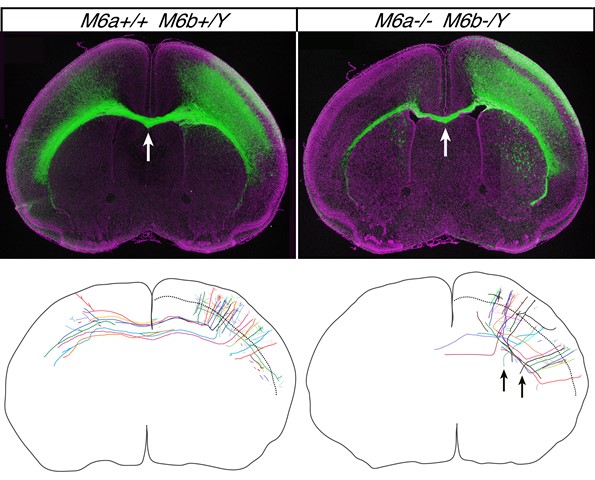
Top: the bundle of callosal axons (green) connecting brain hemispheres (arrow) is thinner in the M6a/M6b double mutant brain (right). Bottom: callosal axon trajectories traced with Supernova system. In the double mutant brain (right), the axons are not simply shorter but also disorganized, and some are even misdirected subcortically (arrows).
Different genetic bases on different aspects of aggression
Mouse Genomics Resource Laboratory (MGRL) • Koide Group
Genetic mapping of escalated aggression in wild-derived mouse strain MSM/Ms: association with serotonin-related genes
Aki Takahashi, Toshihiko Shiroishi, Tsuyoshi Koide Frontiers in Neuroscience Front. Neurosci., 11 June 2014; doi:10.3389/fnins.2014.00156The Japanese wild-derived mouse strain MSM/Ms (MSM) retains a wide range of traits related to behavioral wildness, including high levels of emotionality and avoidance of humans. In this study, we observed that MSM showed a markedly higher level of aggression than the standard laboratory strain C57BL/6J. We identified two chromosomes, Chr 4 and Chr 15, which were involved in the heightened aggression observed in MSM. These chromosomes had different effects on aggression: whereas MSM Chr 15 increased agitation and initiation of aggressive events, MSM Chr 4 induced a maladaptive level of aggressive behavior. Expression analysis of mRNAs of serotonin related genes indicated that the expression of Tph2, an enzyme involved in serotonin synthesis, in the midbrain was increased in the Chr 4 consomic strain, as well as in MSM, and that there was a strong positive genetic correlation between aggressive behavior and Tph2 expression at the mRNA level.
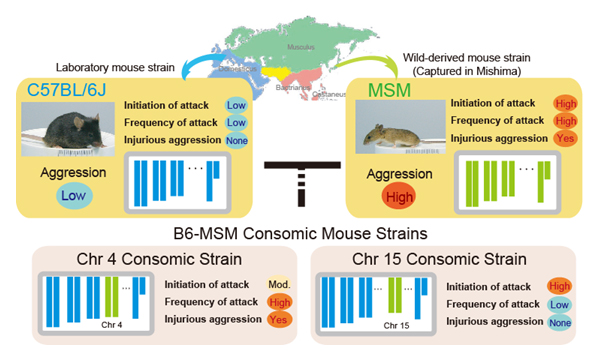
Genetic mapping of escalated aggression in MSM by using consomic mouse strains
Turtle’s and chicken’s brains are similar!?
Division of Brain Function • Hirata Group
A common developmental plan for neocortical gene-expressing neurons in the pallium of the domestic chickenGallus gallus domesticus and the Chinese softshell turtle Pelodiscus sinensis.
I. K. Suzuki and T. Hirata Front. Neuroanat.(2014) 8 20 doi: 10.3389/fnana.2014.00020The highly evolved neocortex is a marked characteristic of mammals. It develops as layers, in which neurons are sequentially added from deep to the surface in a birth-order-dependent manner. Evolutionary processes of the layered neocortex have been a long mystery. We recently reported that the chicken brain contains neuron subtypes that are similar to deep and upper layer neurons in the mammalian neocortex . However, the comparison of only two distant animal groups was not sufficient to specify potential evolutional changes, and similar analyses of other animal groups have been awaited. In this study, we performed gene expression analyses in the softshell turtle brain, which is often referred to as an ancestral form of amniote brains. The turtle brain contains a single neuronal layer, exhibiting a clear difference from the chicken brain. Our expression analyses of orthologs of neocortical layer marker genes highlighted similarities in neuronal arrangements between the two species. Specifically, in both of the species, deep layer neurons are positioned in the medial part, whereas upper layer neurons are positioned in the lateral part. Thus, this medio-lateral neuronal arrangement appears to be an ancestral mode in sauropsids. The results support our hypothesis of an ancient origin of neocortical neuron subtypes. We assume that the generality of birth-order-dependent mechanisms for specification of neuron subtypes probably underlies the their unexpected conservation among diverse amniote species.
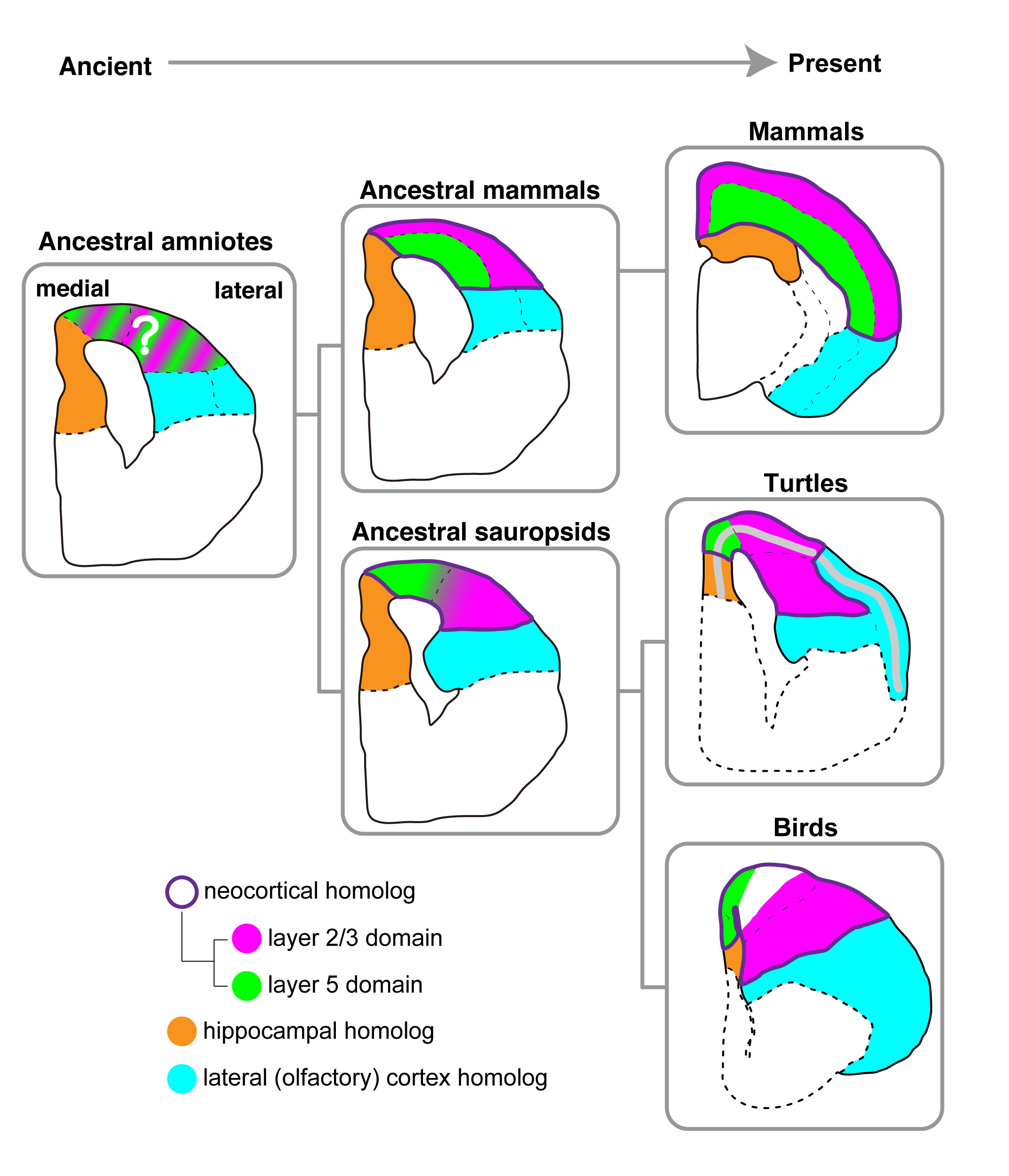
Evolutionary model of brains in amniotes
Development of a freeware that enables automatic characterization of social behavior in mice
Press release
A male-specific QTL for social interaction behavior in mice mapped with automated pattern detection by a hidden Markov model incorporated into newly developed freeware
Toshiya Arakawa1, Akira Tanave1, Shiho Ikeuchi, Aki Takahashi, Satoshi Kakihara, Shingo Kimura, Hiroki Sugimoto, Nobuhiko Asada, Toshihiko Shiroishi, Kazuya Tomihara, Takashi Tsuchiya, Tsuyoshi Koide. ※1 equally contributed.
Journal of Neuroscience Methods Available online 21 April 2014 doi:10.1016/j.jneumeth.2014.04.012
Notwithstanding the importance of effective approaches to analyze social interaction between animals in experimental settings, the methods that are currently available to do this rely predominantly on human observation. This makes large-scale studies of social interaction behavior difficult.
In this paper, we report the development of freeware, called DuoMouse, that enables video recording to track the movements of two mice from a movie file, analysis of behavioral states using a hidden Markov model (HMM), and visualization of the results. We used this software to compare social behavior in consomic and subconsomic strains, and to map a genetic locus responsible for differences in social interaction between the strains. We report a locus that increases social interaction only in males and that lies within a 24.6-Mb region of chromosome 6.
We propose that the method applied in the present study is very efficient and useful for large-scale genetic and pharmacological studies of social behavior in mice. All of the software is open-source, and the source code will also be provided for further development by others.
(https://zenodo.org/records/12577797)
This research was supported by the Research Organization of Information and Systems, Transdisciplinary Research Integration Center, JSPS KAKENHI Grant Numbers 23650243 and 25116527, and NIG Cooperative Research Program (2010-A40, 2012-A85).
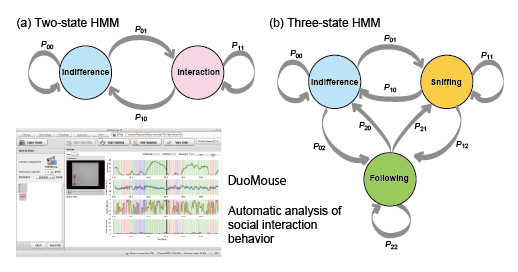
Diagram showing the Markov transition probabilities for the two-state HMM and three-state HMM. The image showing one of the pages of the freeware, DuoMouse.
Development of the lateral line canal system through a bone remodeling process in zebrafish
Division of Molecular and Developmental Biology • Kawakami Group
Development of the lateral line canal system through a bone remodeling process in zebrafish.
Hironori Wada, Miki Iwasaki, Koichi Kawakami Dev. Biol., in press, doi: 10.1016/j.ydbio.2014.05.004The lateral line system of teleost fish is composed of mechanosensory receptors (neuromasts), some of which are superficial whereas others are embedded in canals running under the skin. Canal diameter and size of canal neuromasts are correlated with increasing body size, thus providing a very simple system to investigate the mechanism underlying the coordination between organ growth and body size. Here, we examine the development of the trunk lateral line canal system in zebrafish. We demonstrate that trunk canals originate from scales through a bone remodeling process. We suggest that the bone remodeling process is essential for normal growth of canals and of canal neuromasts. Moreover, we show that the presence of lateral line cells is required for the formation of canals, suggesting the existence of mutual interactions between the sensory system and surrounding connective tissues.
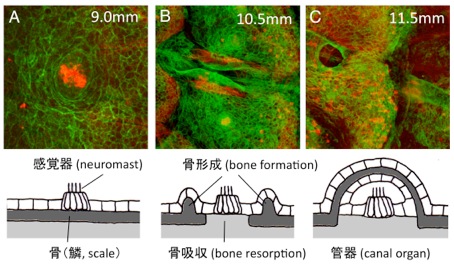
(A) The lateral line sense organ, the neuromast, is located on the skin surface in embryonic stages. (B) When fish undergo metamorphosis, a pair of ridges extends outward from the scale. At the same time, bony elements under the neuromast gradually disappear. (C) The neuromast becomes embedded in the canal running under the scale in adult fish.
Mudi•‘one-click’ identification of causative on web.
Genome Informatics Laboratory • Nakamura Group Division of Cytogenetics • Kobayashi Group
Mudi, a web tool for identifying mutations by bioinformatics analysis of whole-genome sequence.
Naoko Iida, Fumiaki Yamao, Yasukazu Nakamura, and Tetsushi Iida Genes to Cells 28 APR 2014 DOI:10.1111/gtc.12151Genetics is a powerful approach to discovery of the molecular mechanisms underlying biological phenomena. However, experiments for identification of mutations by classical genetics is a time-consuming and laborious process. Modern whole-genome sequencing, coupled with bioinformatics analysis, has enabled fast and cost-effective mutation identification. However, for many experimental researchers, bioinformatics analysis is still a difficult aspect of whole-genome sequencing. To address this issue, we developed a browser-accessible and easy-to-use bioinformatics tool called Mutation discovery (Mudi; http://naoii.nig.ac.jp/mudi_top.html). Users can run Mudi analysis with ‘one-click’ operation in a web browser after uploading genome resequencing data. Mudi simplifies analysis of whole-genome sequencing and thereby expands the possibilities for systematic forward-genetic approaches in various organisms.
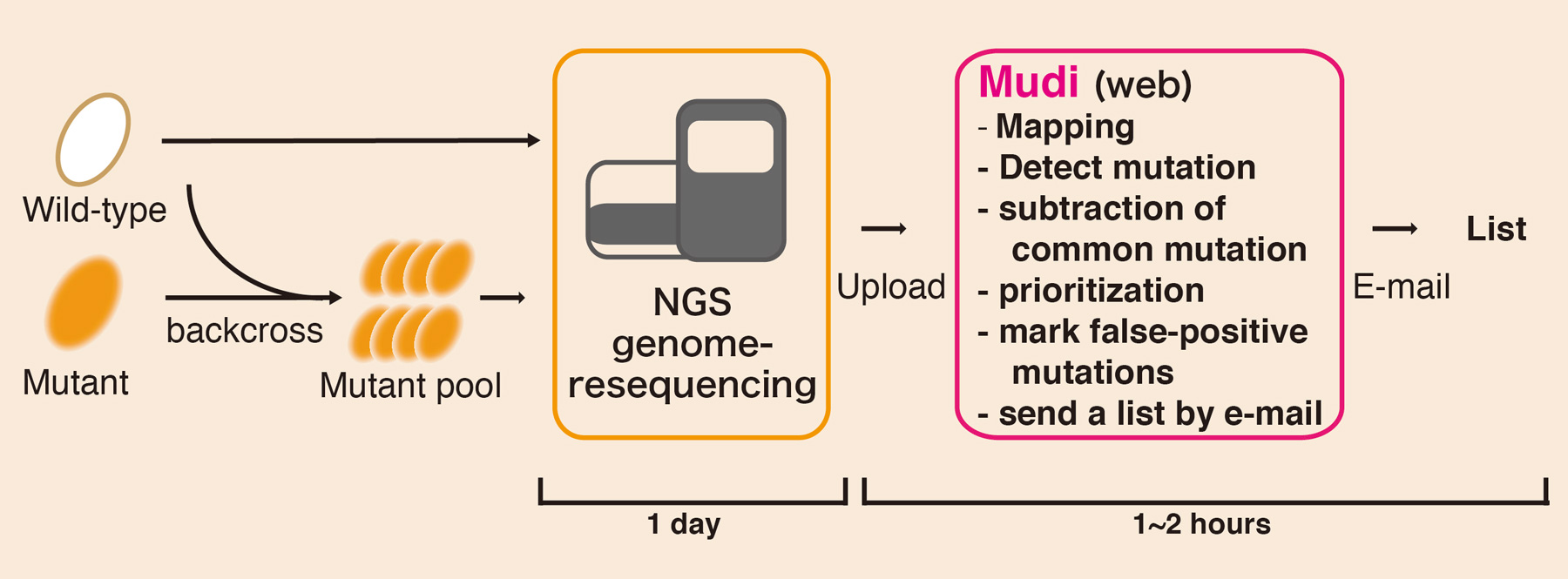
Workflow of “Mudi” system. Genomic DNA is prepared from pooled mutants by pooled-linkage analysis. Mudi calls prioritized mutation candidates by bioinformatics analysis using genome-resequencing data.
A cluster of four QTLs for behavior segregated by new method using a statistical regression model
Mouse Genomics Resource Laboratory (MGRL) • Koide Group
Segregation of a QTL cluster for home-cage activity using a new mapping method based on regression analysis of congenic mouse strains
Shogo Kato, Ayako Ishii, Akinori Nishi, Satoshi Kuriki, Tsuyoshi Koide Heredity. advance online publication 30 April 2014; doi:10.1038/hdy.2014.42Genetic mapping using congenic strains is one of the most powerful methods available to show the existence of QTLs in the genomic regions substituted into recipient strains. In this case, genetic mapping can be conducted by making a series of congenic strains that cover genomic regions that partially overlap with the adjacent congenic strains. In the analysis of the phenotype using a series of congenic strains, however, the phenotype shows a variety of levels, which indicates the existence of multiple QTLs in the mapped chromosomal regions. These results show that the precise genetic mapping of fine QTLs is extremely difficult using current methods.
Dr. Tsuyoshi Koide’s group collaborated with Drs. Shogo Kato and Satoshi Kuriki in the Institute of Statistical Mathematics and established a new method for mapping multiple QTLs using data from a series of congenic strains by applying a regression model for the analysis of total home-cage activity in mice. The results of the analysis identified four significant QTLs in a 14.5 Mb genomic region. Among these, three have negative effects but one has a positive effect on total home-cage activity. In further analysis of recombinants obtained from the congenic strains, we confirmed the existence of the QTL that has a positive effect on the activity as well as another QTL that suppresses the effect of this positive QTL. These results clarify for the first time the association of a complex genetic mechanism in the QTL cluster on chromosome 6 with the regulation of home-cage activity.
This work was supported by the Research Organization of Information and Systems, Transdisciplinary Research Integration Center, JSPS KAKENHI (Grant Numbers 23650243 and 25116527), and Yamada Science Foundation.
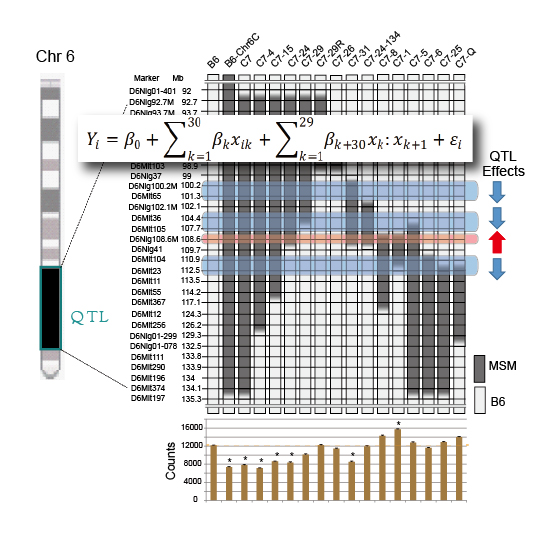
A cluster of four QTLs for behavior segregated by new method using a statistical regression model. The data of genomic regions carried in the congenic strains as well as the data of home-cage activity exhibited by congenic strains are analyzed with regression model. The study revealed four QTLs which are clustered in a small genomic region.
Ms. Neha Mishra (D3 Student SOKENDAI at Division of Evolutionary Genetics)received SOKENDAI President’s Award.

X chromosome-autosomes incompatibility accelerates speciation: reproductive isolation by misregulation of X-linked genes
Press release
Evolutionarily Diverged Regulation of X-chromosomal Genes as a Primal Event in Mouse Reproductive Isolation
Ayako Oka, Toyoyuki Takada, Hironori Fujisawa, and Toshihiko Shiroishi PLoS Genet Accepted manuscript online, 2014, doi/10.1371/journal.pgen.1004301Reproductive isolation is important for speciation, because it suppresses free genetic exchange between two diverged populations and accelerates the genetic divergence. Hybrid sterility (sterility in hybrid animals), one of the reproductive isolation phenomena, is possibly caused by deleterious interactions between diverged genetic factors brought by two distinct populations. A mouse inter-subspecific X chromosome substitution strain, B6-ChrXMSM, in which X chromosome of west European subspecies-derived C57BL/6J is replaced by counterpart of Japanese subspecies-derived MSM/Ms strain, shows reproductive isolation characterized by male-specific sterility due to disruption of meiotic entry in spermatogenesis (Oka et al, Genetics, 2004, 2007, 2010). In this study, we conducted comprehensive transcriptional profiling of the testicular cells of this strain by microarray. The results clearly revealed gross downregulation of gene expression in the substituted donor X chromosome. Such misregulation occurred prior to detectable spermatogenetic impairment, suggesting that it is a primal event in reproductive isolation. Furthermore, this misregulation subsequently resulted in perturbation of global transcriptional regulation of autosomal genes, probably by cascading deleterious effects. Remarkably, this transcriptional misregulation was substantially restored by introduction of chromosome 1 from the same donor strain as the X chromosome. This finding implies that one of regulatory genes acting in trans for X-linked target genes is located on chromosome 1. This study collectively suggests that regulatory incompatibility is a major cause of reproductive isolation in the X chromosome substitution strain.
This study was done as a research project “Life Systems” of Trans- disciplinary Research Integration Center, ROIS.
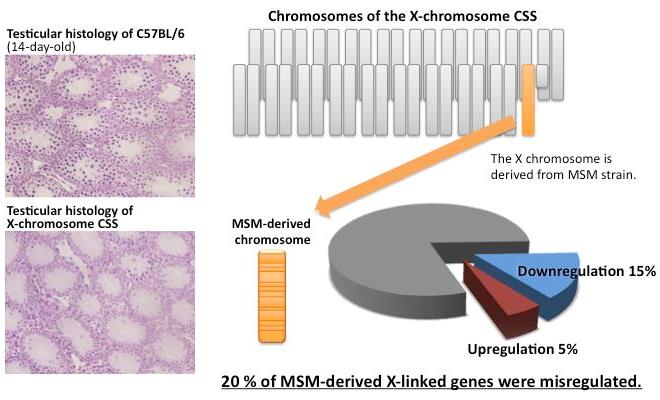
Figure 1. Misregulation of X-linked genes in the X-chromosome substitution strain (CSS). Meiotic spermatocytes were observed in testes of wild type (left upper panel), whereas they were rarely observed in testes of X-chromosome CSS (left bottom panel). Approximately 20% of expressed X-linked genes were misregulated in the X-chromosome CSS.
The inhibitory role of the medial prefrontal cortex on aggressive behavior in male mice
Press release
Control of Intermale Aggression by Medial Prefrontal Cortex Activation in the Mouse.
Aki Takahashi, Kazuki Nagayasu, Naoya Nishitani, Shuji Kaneko, Tsuyoshi Koide PLOS ONE April 16, 2014 doi:10.1371/journal.pone.0094657The adaptive nature of aggressive behavior in social animals accounts for its observation throughout the animal kingdom. However, given that aggressive behavior is no longer adaptive when it exceeds the species-typical level, mechanisms should curb excessive aggression to keep it within the adaptive range. The prefrontal cortex has been implicated in the inhibitory control of emotional outbursts, including aggression and violence. In this study, we used optogenetics to demonstrate that temporal activation of excitatory neurons in the medial prefrontal cortex (mPFC), but not the orbitofrontal cortex (OFC), inhibits inter-male aggression in mice. Activation of the mPFC suppresses aggressive bursts and reduces the intensity of aggressive behavior, but does not change the duration of the aggressive bursts. Our findings suggest that mPFC activity inhibits the initiation and execution, but not the termination, of aggressive behavior, and maintains such behavior within the adaptive range.
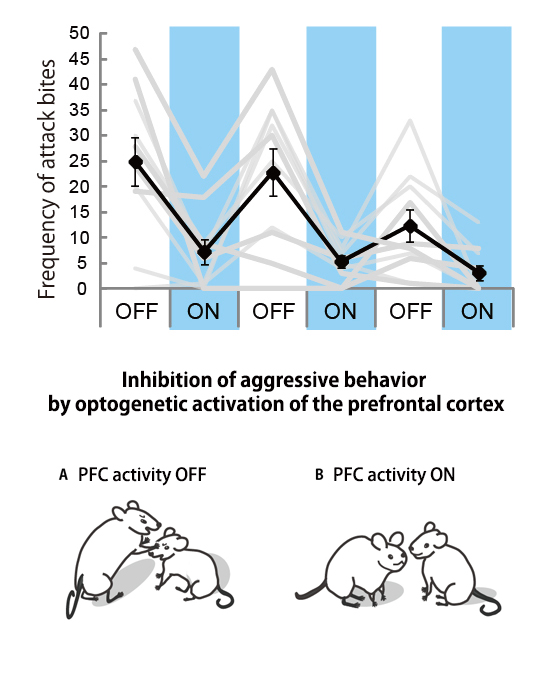
Optogenetic activation of the mPFC reduced the frequency of attack bites.
ON: light stimulation (blue light, 20 Hz, 5 mW), OFF: no light stimulation, gray lines: attack bites in each individual, black line: mean values.
Hideaki Kanzawa receives “Morishima Award”
 Hideaki Kanzawa, Ph.D. candidate at the Division of Population Genetics, received “Morishima Award for outstanding doctoral research in the Department of Genetics, SOKENDAI”. The awarding ceremony was held on 11th March, 2014.
Hideaki Kanzawa, Ph.D. candidate at the Division of Population Genetics, received “Morishima Award for outstanding doctoral research in the Department of Genetics, SOKENDAI”. The awarding ceremony was held on 11th March, 2014.Morishima Award
About Dr. Hiroko Morishima (“Pioneers in Genetics” vol.3)
Small RNA biogenesis specific to reproductive tissues in rice
Experimental Farm • Nonomura Group Plant Genetics Laboratory • Kurata Group
Rice Germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs
Reina Komiya, Hajime Ohyanagi, Mitsuru Niihama, Toshiaki Watanabe, Mutsuko Nakano, Nori Kurata, and Ken-Ichi Nonomura The Plant Journal Accepted manuscript online, 2014, doi:10.1111/tpj.12483Meiosis is a pivotal biological event for breeding programs, and elucidatiing molecular mechanisms of meiosis will lead to achieve stable and efficient seed production. In this paper, we focus on MEL1 protein, which plays essential roles in progression of meiosis in rice. MEL1 is an Argonaute family protein known as an effecter of RNA silencing.
Of all MEL1-binding small RNAs (MEL1-sRNA) isolated and sequenced, 75% were 21 nt long and 80% beared a 5′-terminal cytosine. Supprisingly, MEL1-sRNA derived from more than 1,000 genomic loci, and most of the loci were mapped onto the intergenic regions. We discovered that more than 700 species of large intergenic noncoding RNAs (lincRNA) were transcribed from the MEL1-sRNA loci after the reproductive phase transition. The lincRNAs were double-stranded and processed in the 21-nt phase by tasiRNA biogenesis pathway, resulting in production of the 21-nt small RNAs including MEL1-sRNA.
This study brought us important information to consider the role of RNA silencing in plant germline development and meiosis.
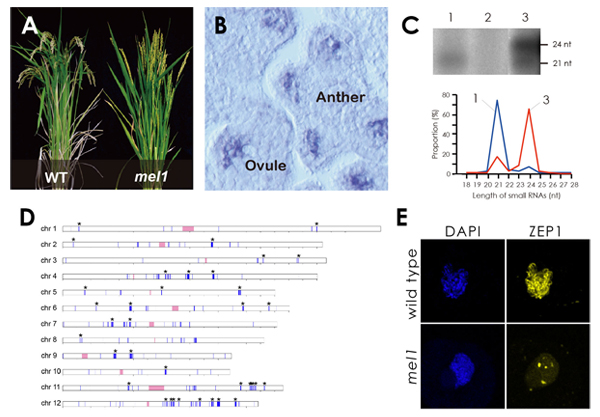
Analysis of MEL1, the rice Argonaute protein with specific expression in the germline.
(A) The mel1 mutant is sterile. (B) MEL1 gene is expressed in germ cells. (C) RNA-immunoprecipitation of wt (1) and muntat panicles (2). (D) MEL1-binding small RNAs derive from >1,000 intergenic loci (blue). (E) ZEP1 doesn’t elongate between homolog pairs in mel1 meiocytes.
Panels A-B and C-E were cited from Plant Cell 19: 2583-2594 (2007) and Plant J (2014), respectively, with slight modifications.















