Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
How Life Thrives in a Nutrient-Poor Environments — “Circular Agriculture” Inside Cells through Photosynthesis and Symbiosis
Miyagishima Group / Symbiosis and Cell Evolution Laboratory
The closed nutrient recycling system in the Paramecium–Chlorella photosymbiosis contributes to survival under oligotrophic conditions
Kaoru Okada, Takayuki Fujiwara, Shunsuke Hirooka, Yusuke Kobayashi, Ryo Onuma, and Shin-ya Miyagishima
Science Advances 11, eadz0004 (2025) DOI:10.1126/sciadv.adz0004
Endosymbiotic relationships between a heterotrophic host and a unicellular algal endosymbiont are observed across many eukaryotic lineages. Although these relationships are prevalent in oligotrophic environments, how they function and provide an advantage under such conditions remains largely unknown. To address these issues, we examined the behaviour of the ciliate Paramecium bursaria hosting Chlorella endosymbionts under nitrogen- and prey-depleted conditions. The Paramecium host survived for up to five weeks while maintaining the number of Chlorella endosymbionts, whereas aposymbiotic Paramecium and free-living Chlorella either died or bleached, respectively, under the same conditions. In the symbiotic state, the host continuously fed on the endosymbionts without excreting nitrogenous waste into the medium, while the remaining endosymbionts continued to proliferate using heterotrophic metabolites from the host and light energy. Thus, the cyclical farming of endosymbionts by the host maintains a high concentration of nutrients within the closed system, providing a selective advantage in oligotrophic environments.
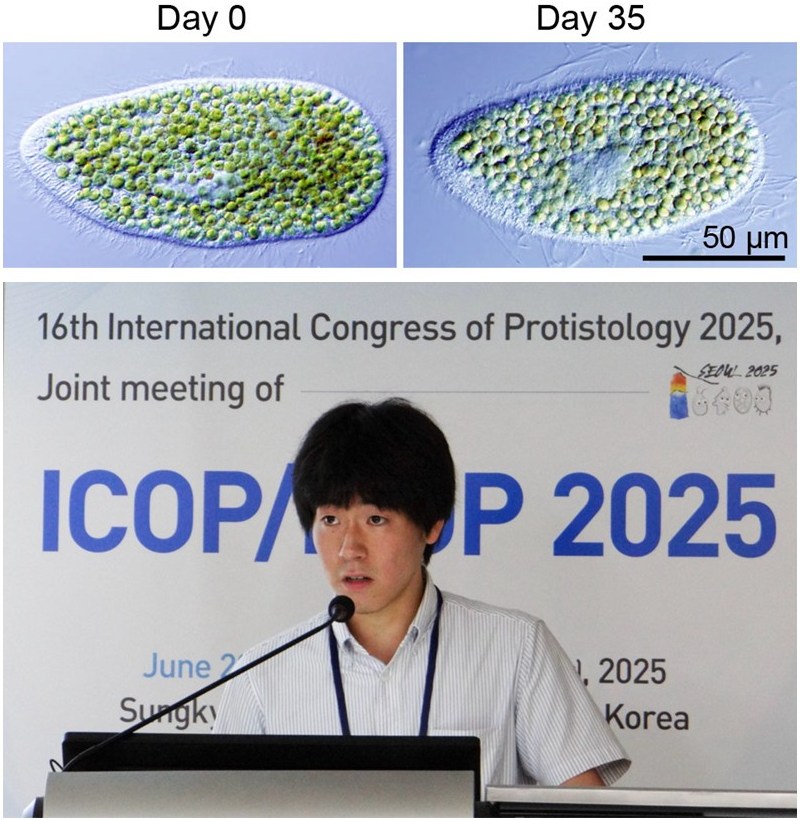
Figure. Micrographs of Paramecium bursaria and Okada-san (a graduate student) presenting the findings at an international conference
A Paramecium bursaria cell (Day 0) that had been grown with bacterial prey was further incubated for 35 days under light in a culture medium lacking prey as well as nitrogen and phosphorus sources (Day 35). The host paramecium remained viable while maintaining approximately 300 endosymbiotic Chlorella cells per host cell. These Chlorella cells were partly digested by the host but also proliferated, resulting in the apparent number of symbionts being largely maintained. Okada-san presented these findings at ICOP/ISOP 2025, an international protozoology conference held in Seoul, Korea, from June 22 to 27, 2025, and received the Best Oral Presentation Award
Why does ALS take away body movement? – The hidden burden that seals neurons’ fate
Press release
Intrinsically accelerated cellular degradation is amplified by TDP-43 loss in ALS-vulnerable motor neurons in a zebrafish model
Kazuhide Asakawa, Takuya Tomita, Shinobu Shioya, Hiroshi Handa, Yasushi Saeki, and Koichi Kawakami
Nature Communications DOI:10.1038/s41467-025-65097-0
![]() Press release (In Japanese only)
Press release (In Japanese only)
Amyotrophic lateral sclerosis (ALS) is a fatal disease in which motor neurons that control voluntary movement gradually degenerate, leading to paralysis. Although many studies have been conducted, the reason why motor neurons are selectively affected has remained unclear.
Using transparent zebrafish that allow live imaging of nerve cells, we found that motor neurons constantly endure a burden to keep proteins properly folded (Figure 1). Larger neurons were particularly vulnerable, and this burden was further increased by genetic manipulations mimicking ALS.
Because motor neurons connect distant organs—brain, spinal cord, and muscles—with single cells, they must grow large and synthesize vast amounts of proteins, which increases their burden. This “inevitable degradation burden” may underlie their selective vulnerability in ALS. Even in tiny zebrafish larvae, such burden was detected, suggesting that in humans, whose motor neurons can reach as long as one meter in length, the challenge is far greater.
Understanding how this degradation burden arises may open the way to new therapeutic strategies to slow ALS progression and protect motor neurons from degeneration.
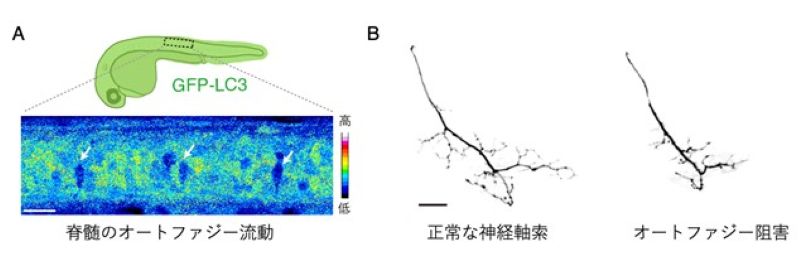
Figure: A) Large motor neurons show high protein degradation activity (autophagic flux). GFP-LC3 is degraded through autophagy. Arrows indicate large motor neurons with active GFP-LC3 degradation.
B) Inhibition of autophagy impairs axonal development of motor neurons (right). Scale bars, 20 µm.
Single-Strand DNA Cuts Trigger Cancer—Mechanism Behind Copy Number Abnormalities Discovered
Press release
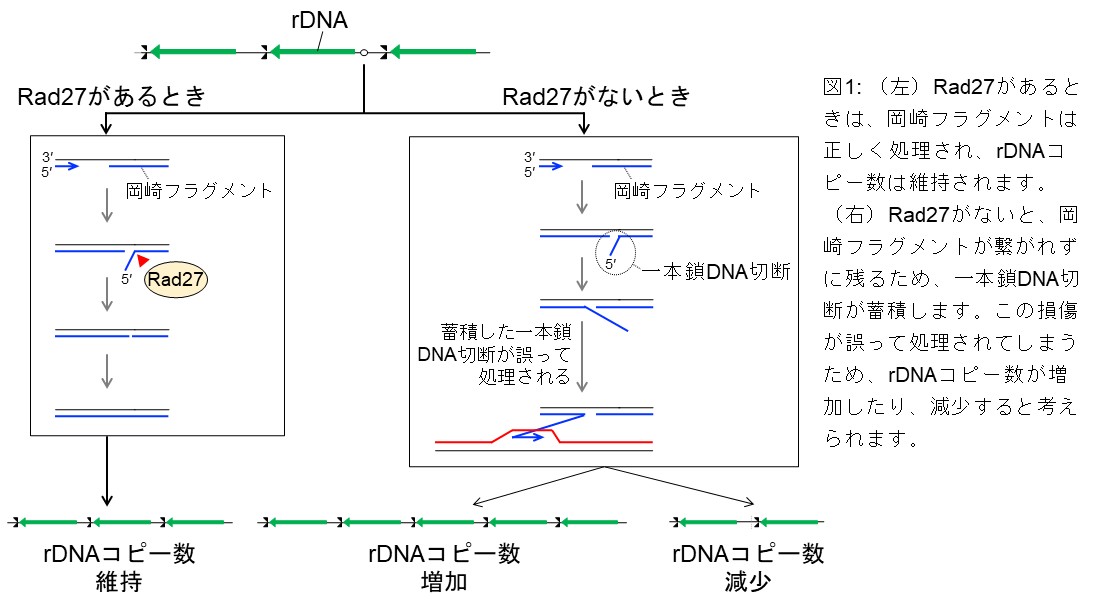
Rad27/FEN1 prevents accumulation of Okazaki fragments and ribosomal DNA copy number changes
Tsugumi Yamaji, Yuko Katayama, Nanase Arata, and Mariko Sasaki
FEBS Letters 2025 DOI:10.1002/1873-3468.70193
![]() Press release (In Japanese only)
Press release (In Japanese only)
DNA copy number changes are the most frequent genomic alterations in cancer cells. Here, we 19 demonstrate that Rad27/FEN-1, a structure-specific nuclease in budding yeast, plays a crucial role in 20 maintaining the stability of the ribosomal DNA (rDNA) repeats. Severe rDNA instability is observed 21 in the rad27∆ mutant, independently of Fob1-mediated DNA replication fork arrest and DNA double-22 strand break (DSB) formation in the rDNA. The rad27Δ mutant accumulates Okazaki fragments in the 23 rDNA region, without inducing formation of detectable DSBs. Similar rDNA instability is observed in 24 DNA ligaseCdc9-deficient cells. Furthermore, Exonuclease 1 and PCNA partially compensate for the 25 loss of Rad27 in the rDNA stabilization. These findings highlight the importance of proper Okazaki 26 fragment maturation in the maintenance of rDNA stability.
Full /Associate Professor (Department of Gene Function and Phenomics)
The National Institute of Genetics (NIG) invites applications for one full or associate professor position in the Department of Gene Function and Phenomics. Please see the details below and apply accordingly.
Application Guidelines:PDF Application Materials:Brief summary of career(excel)
1. Division: Department of Gene Function and Phenomics
2. Recruiting: Full /Associate Professor (1 qualified person)
3. Appointment Start Date: On and after April 2026 (negotiable)
4. Term of Appointment:
Indefinite term appointment (until the mandatory retirement age of 65, based on the regulations of the Research Organization of Information and Systems)
5. Qualifications:
The candidate should be a talented researcher with outstanding achievements in microbiology or any other fields of life science related to microorganisms, have the expertise and leadership skills necessary to conduct cutting-edge research in these fields, and develop the prokaryote bioresource at the National Institute of Genetics.
6. Responsibilities:
The appointed researcher will lead an independent laboratory as a principal investigator. He or she will also participate in the education of graduate students as a faculty member of the Genetics Program of SOKENDAI.
7. Research environment and support:
・As an Inter-University Research Institute, NIG provides substantial common equipment and research infrastructure and promotes collaborative research worldwide. We expect the appointed researcher to take advantage of this research environment to organize and lead his/her research team.
・One assistant professor and one technical support staff member will be allocated to the laboratory.
8. Application Deadline: Noon (Japan Time) on December 24, 2025
9. Application Materials:
I. Curriculum Vitae (In English. Japanese applicants should also submit a Japanese version. Include your e-mail address.)
II. List of publications (If there are multiple authors, briefly describe your contribution. Indicate your most significant papers.)
III. Summary of past and present research and future directions (In English, max. 1,500 words. Include figures if necessary.)
IV. Names and contacts of references (At least two domestic and two international references.)
V. Summary of career
VI. Your key papers
*All the personal data in the application is handled in strict confidentiality, and used solely for recruitment purposes.
10. Submission:
Submit items I to VI listed above electronically as follows:
(a) The subject line should be “Application for Department of Gene Function and Phenomics Full / Associate Professor”, which should be also noted in the e-mail body.
(b) Put items I to IV together in a single file by separating each item by page. E-mail the file as an attachment. The file format should be MS Word or PDF. Also attach item V (form can be downloaded from the NIG homepage) as a separate file.
(c) Send your key papers as PDF attachments. The file format should be PDF. If your key papers are available to access online, include a list of URLs in the e-mail body.
* We will notify you via e-mail upon receipt of your electronic application within two business days.
Based on gender equality National Institute of Genetics has been actively promoting female scientists. In case of equivalent aptitude and achievements in research, education, and social contributions, preference will be given to female candidates.
National Institute of Genetics is promoting safety and healthy working, including preventing passive smoking. (Indoor smoking is banned. / Smoking is allowed only in the designated outdoor areas.)
Send your application and inquiries to:
NIG Personnel Committee (Personnel Team)
E-mail:
Mailing Address:
National Institute of Genetics
Yata 1111, Mishima, Shizuoka 411-8540 JAPAN
TEL: +81-55-981-6716
Homepage: https://www.nig.ac.jp/nig/
NIG Organization Chart: https://www.nig.ac.jp/nig/research/organization-top/organization
[Notes on Submission of Documents]
The following deficiencies have been observed:
Cases where the PDF file of the key paper was not attached
Cases where access to the key paper on the internet required an account, thereby restricting viewing
If, due to file size limitations, it is difficult to send all PDF files in a single email, they may be divided across multiple emails without issue.
In any event, please ensure that the PDF files are duly submitted.
Furthermore, if access to a paper requires an account on the relevant website, kindly indicate this in the remarks section.
Dr. Katsuhiro Yoneoka won the JPR Best Paper Award at the Botanical Society of Japan

BSJ President Hasebe (left)
Dr. Katsuhiro Yoneoka, a postdoctoral researcher in the Plant Evolution Laboratory (Fukushima Lab), received the JPR Best Paper Award at the 89th Annual Meeting of the Botanical Society of Japan, held at the Fukuoka International Congress Center, Fukuoka, Japan, on September 17– 20, 2025.
▶ Awarded presentation title: Morphological and functional evolution of gametophytes in epilithic Hymenasplenium murakami-hatanakae (Aspleniaceae): The fifth family capable of producing the independent gametophytes
Dr. Masahito Tanaka wins the Early Career Presentation Award at the 63rd Annual Meeting of the Biophysical Society of Japan

Dr. Masahito Tanaka, from the Laboratory of Physics and Cell Biology (the Shimamoto Lab) at the National Institute of Genetics, gave an invited talk at the 63rd annual meeting of the Biophysical Society of Japan and won the Early Career Presentation Award. Dr. Tanaka has been supported by a JSPS postdoctoral fellowship since 2023 and will start his new research under JST ACT-X this October.
▶ Awarded presentation title: Changes in the physical properties of early embryonic nuclei promote a transcriptional burst
▶ The 63rd Annual Meeting of the Biophysical Society of Japan
Selective breeding makes mice playful towards humans and other mice Tame mice show human- and mouse-directed playfulness
Koide Group / Mouse Genomics Resource Laboratory
Comparative Analysis of Tickling and Conspecific Play in Tame Mice and Golden Hamsters
Dagher S, DeAngelo D, Sato RY, Norimoto H, Koide T*, and Ishiyama S*
(*co-corresponding author)
Behavioural Brain Research (2026) 496 DOI: 10.1016/j.bbr.2025.115849
Social “play” is a vital behaviour through which animals strengthen bonds, acquire essential skills, and express positive emotions. Yet mice have long been regarded as creatures that simply do not play.
A research team led by Dr. Sarah Dagher (PhD student at the time of the experiment, now at the Max Planck Institute for Metabolism Research) and Dr. Shimpei Ishiyama (now at the Central Institute of Mental Health, Mannheim), and Associate Professor Tsuyoshi Koide at the National Institute of Genetics has now overturned this long-held belief. They discovered that “tame mice” — selectively bred to approach a human hand — display playful behaviours with both humans and fellow mice.
When tickled, these tame mice emit ultrasonic sounds resembling laughter and actively chase the researcher’s hand. In social settings, they engage in playful behaviours such as poking at one another and making exaggerated movements, again accompanied by ultrasonic vocalisations. By contrast, ordinary mice show little response to human tickling, interact less with peers, and often display aggression instead.
Intriguingly, the tame mice produce different types of vocalisations when playing with humans compared to when playing with other mice, indicating that they clearly distinguish their social partners.
These findings have important implications for understanding animal domestication and human–animal relationships. Breeding mice for tameness not only increased their willingness to play with humans but also enhanced social play among peers. Previous genetic analyses further revealed that regions associated with tameness in mice overlap with those linked to domestication in dogs, suggesting a shared mechanism across species.
This study shows that a tendency to “enjoy play” is not limited to certain animals but can emerge as a result of increased receptiveness to human interaction. It also challenges the long-standing notion that “rats play but mice do not,” demonstrating that mice, too, can become playful companions — both for humans and for one another.
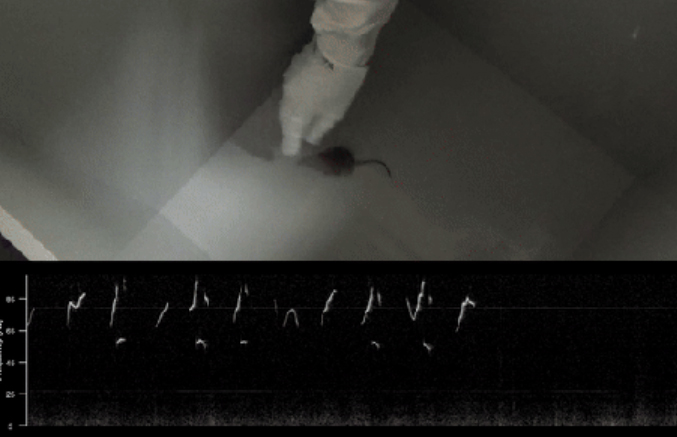
Figure: The selectively bred mice respond to tickling by emitting ultrasonic vocalisations and exhibiting play-like behaviour, such as spontaneously chasing the hand.
The video is here: https://neurogelotology.lol/2025/09/30/selective-breeding-makes-mice-playful-towards-humans-and-other-mice/
“Dean’s Award” awarded to Genetics Program student Islam, Moutushi
Ms. Islam, Moutushi, who graduated from SOKENDAI in September 2025 as a member of the Molecular Cell Engineering Laboratory, has been awarded the School of Life Science “Dean’s Award” for the first semester of 2025.
The Dean’s Award recognizes degree recipients who have conducted research worthy of commendation and reported their accomplishments in an outstanding doctoral thesis. The award was presented on September 26, 2025 during the graduation ceremony. In addition to the Dean’s Award, Ms. Islam has also been awarded the Genetics Program’s Morishima Award.
・Thesis title : Advancing the utility of AID2-based conditional protein knockdown
Ms. Islam has provided the following statement regarding the award.
“I am honored and delighted to receive the Dean’s Award from the School of Life Science, SOKENDAI University.
I would like to express my heartfelt gratitude to my supervisor, Professor Masato Kanemaki. Without his constant guidance and support, this achievement would not have been possible.
National Institute of Genetics, Japan is the first place where I truly learned about research, and it has provided me with invaluable opportunities to explore and grow in this field. I am also grateful to my lab members and collaborators for their contributions, and to my family for their continuous encouragement throughout this journey.
This recognition will inspire me to continue pursuing science with passion and dedication. It will remain a strong motivation as I move forward in my career.”

Ms. Islam
Wild animal populations carrying mutations similar to those that cause human diseases
Kitano Group / Ecological Genetics Laboratory
Functional mutations in the thyroid-stimulating hormone receptor in natural stickleback populations at sites identical to human disease-causing mutations
Jun Kitano, Mana Sato, Hiyu Kanbe, Genta Okude, Asano Ishikawa, Yukinori Kazeto & Takashi Makino
BMC Ecology and Evolution (2025) 98 DOI:10.1186/s12862-025-02440-5
This study examined whether mutations that cause thyroid disease in humans could help identify functional mutations in natural stickleback populations. The researchers found that several Japanese stickleback populations carry non-synonymous mutations in the thyroid-stimulating hormone receptor (Tshr)2 gene. Functional assays revealed that amino acid substitutions at sites corresponding to human loss-of-function or gain-of-function mutations similarly reduced or enhanced receptor activity. Thus, we discovered natural populations carrying mutations similar to those that cause human disease. Further research is needed to determine whether these mutations are deleterious or adaptive in their habitats.

Figure: Hariyo (a freshwater population of the genus Gasterosteus, currently found in Gifu and Shiga Prefectures, and formerly inhabiting Mie Prefecture, Japan) possessed a mutation in the thyroid-stimulating hormone receptor (TSHR) gene at an identical site where mutations cause human disease.
Photo courtesy of Yasuyuki Hata.















