Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
The TEDC1 mutation cause severe growth impairment in humans and zebrafish
Sakai Group / Model Fish Genetics Laboratory
Biallelic TEDC1 variants cause a new syndrome with severe growth impairment and endocrine complications
Noriko Miyake, Kentaro Shiga, Yuya Hasegawa, Chisato Iwabuchi, Kohei Shiroshita, Hiroshi Kobayashi, Keiyo Takubo, Fabien Velilla, Akiteru Maeno, Toshihiro Kawasaki, Yukiko Imai, Noriyoshi Sakai, Tomonori Hirose, Atsushi Fujita, Hidehisa Takahashi, Nobuhiko Okamoto, Mikako Enokizono, Shiho Iwasaki, Syuichi Ito, Naomichi Matsumoto
European Journal of Human Genetics 2025 Feb 20. DOI:10.1038/s41431-025-01802-3
We encountered two affected male patients born to non-consanguineous parents, who presented with prenatal-onset severe growth impairment, primary microcephaly, developmental delay, adrenal insufficiency, congenital glaucoma, delayed bone aging, craniosynostosis, congenital tracheal stenosis, and primary hypogonadism. By exome sequencing, we identified compound heterozygous TEDC1 variants (NM_001134877.1 c.[104-5C>G];[787delG] p.[?];[(Ala263LeufsTer29)] in both affected siblings. We confirmed that the splice site variant, c.104-5C>G leads to no TEDC1 protein production via nonsense-mediated mRNA decay. The frameshift variant located in the last coding exon, c.787delG, produces a C-terminally truncated protein, which impairs the binding with TEDC2. Thus, both variants are thought to be loss-of-function. TEDC1 and TEDC2 are both required for centriole stability and cell proliferation. Our in vitro experiments using patient-derived cells revealed cell cycle abnormality. Our in vivo study using tedc1−/− zebrafish generated by CRISPR/Cas9 successfully recapitulated the growth impairment and cranial bone dysplasia as seen in our patients. The tedc1−/− mutant zebrafish were sterile and did not have developed gonads. Furthermore, we showed that biallelic TEDC1 deletion causes cilia abnormalities through defective acetylated tubulins.
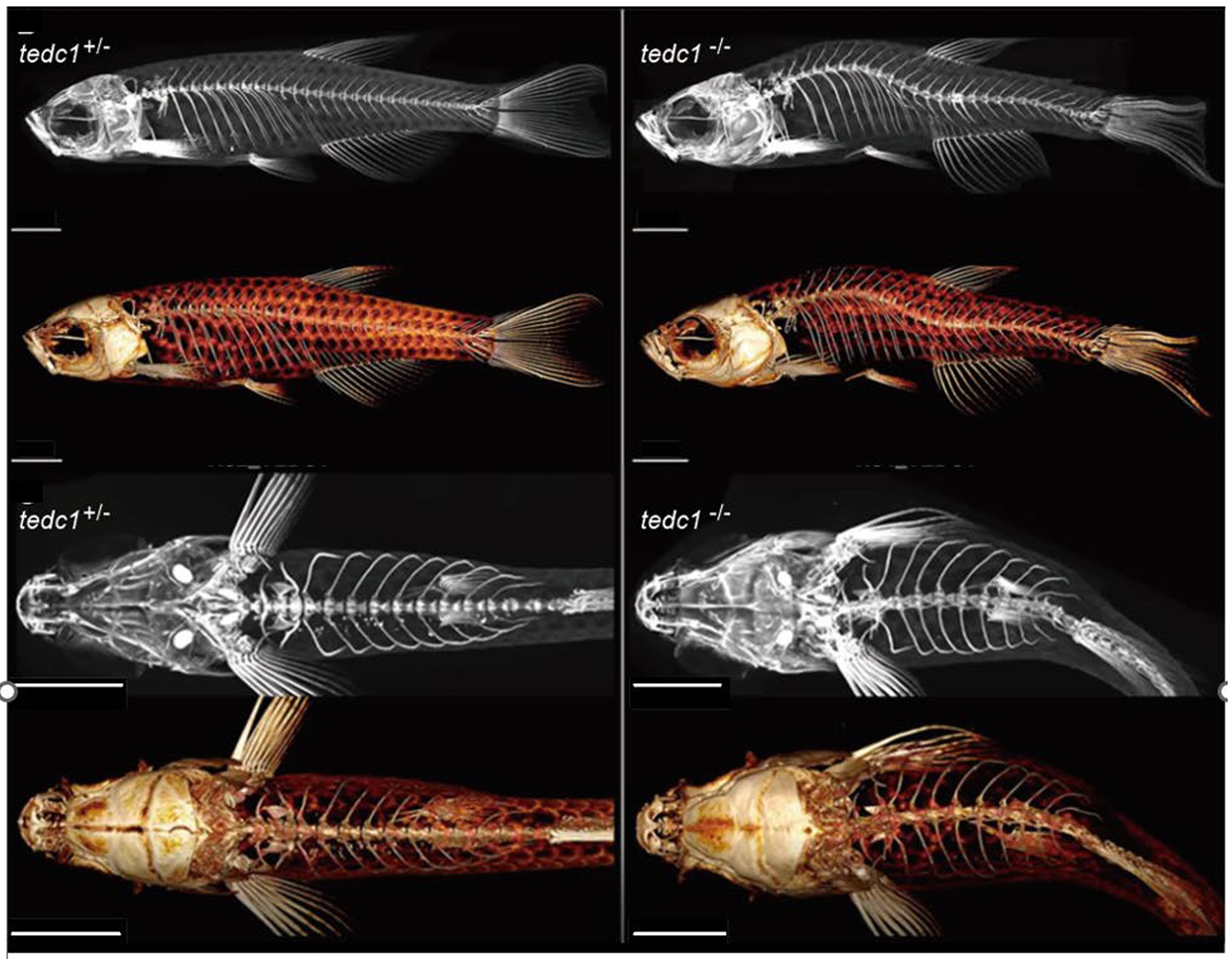
Figure: Representative lateral and upper view µCT images of tedc1+/− and tedc1−/− zebrafish
Chromatin domains in the cell
Maeshima Group / Genome Dynamics Laboratory
Chromatin domains in the cell: phase separation and condensation
Shin Fujishiro*, Masaki Sasai*, and Kazuhiro Maeshima*
*Corresponding author
Current Opinion in Structural Biology Volume 91, April 2025, 103006 DOI:10.1016/j.sbi.2025.103006
Free download link for 50 days [Available online 20 February 2025]
Negatively charged genomic DNA wraps around positively charged core histone octamers to form nucleosomes, which, along with proteins and RNAs, self-organize into chromatin within the nucleus. In eukaryotic cells, chromatin forms loops that collapse into chromatin domains and serve as functional units of the genome. Since various factors—such as chromatin-binding proteins, histone modifications, transcriptional states, depletion attraction, and cations—can significantly impact chromatin organization, the formation processes of these hierarchical structures remain unclear. In this review paper, Shin Fujishiro (Kyoto Univ), Masaki Sasai (Kyoto Univ) and Kazuhiro Maeshima (NIG) critically discuss the formation mechanisms of the chromatin domain in the cell from a physical point of view, including phase separation and condensation.
This work was supported by the JSPS and MEXT KAKENHI grants (JP20H05936, JP22H00406, JP23K17398, and JP24H00061), and the Takeda Science Foundation.
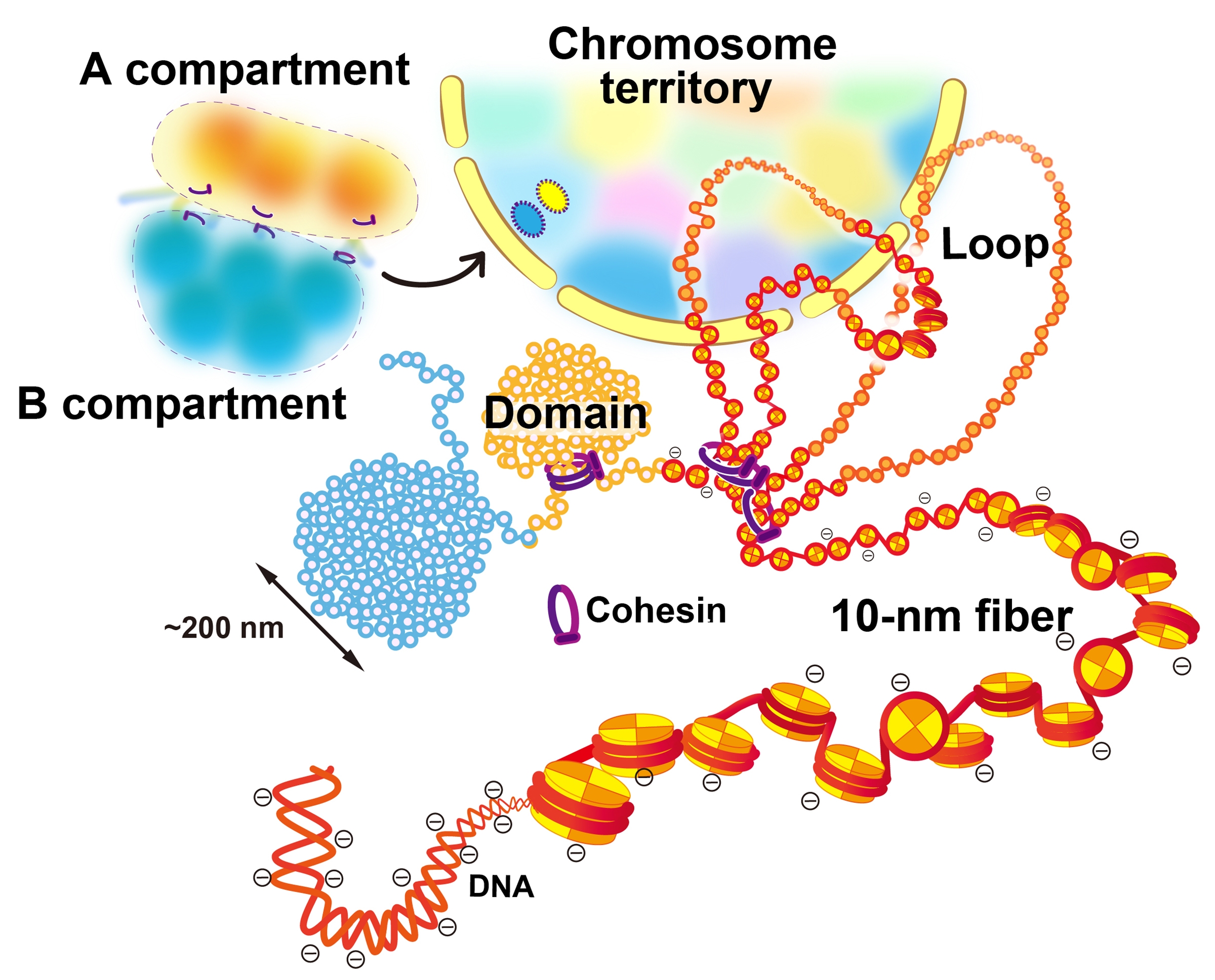
Figure: Scheme for hierarchical chromatin organization inside the cell nucleus.
DNA is wrapped around histone octamer to make a nucleosome. The chain of nucleosomes is compacted into chromatin domains, which interact over long distances to form chromatin compartments. A and B compartments in general represent transcriptionally active or open chromatin state (compartment A) and inactive or closed chromatin (compartment B), respectively. A single interphase chromosome consists of several compartments occupies that collectively form a chromosome territory. Note that this scheme is highly simplified and a more complex organization can be possible inside the cell.
Visualizing the Organized Flow of Cells with 3D Modeling
Press release
Swirling Instability mediated by Elastic and Hydrodynamic Couplings in Cytoplasmic Streaming
Takuji Ishikawa*, Takayuki Torisawa, Hirofumi Wada, Akatsuki Kimura*.
*corresponding authors
PRX Life (2025) 3, 013008 DOI:10.1103/PRXLife.3.013008
![]() Press release (In Japanese only)
Press release (In Japanese only)
Cells contain numerous molecules that collectively function to maintain cellular activity. One striking example is cytoplasmic streaming, which is coordinated intracellular flow. Although individual molecules interact locally, how their movements collectively form an organized flow remains a mystery. In 2017, Prof. Kimura’s team studied cytoplasmic streaming in C. elegans zygotes and discovered that microtubules align via endoplasmic reticulum (ER) connections, and direct flow. At that time, Prof. Joanny (Curie Institute) developed a 1D mathematical model to explain flow initiation and reversal. However, a 3D simulation is required for more detailed comparison with real cells.
In the present study, Prof. Ishikawa (Tohoku Univ.) created a 3D computational model, refined through collaboration with Prof. Wada (Ritsumeikan Univ.). This revealed that ER elasticity is crucial—too weak, flow ceases, too strong, reversals disappear. These results matched the experimental data obtained by Dr. Torisawa (Cell Architecture Lab, NIG). This study quantitatively explains cytoplasmic streaming dynamics and provides a foundation for understanding intracellular flow in various organisms.
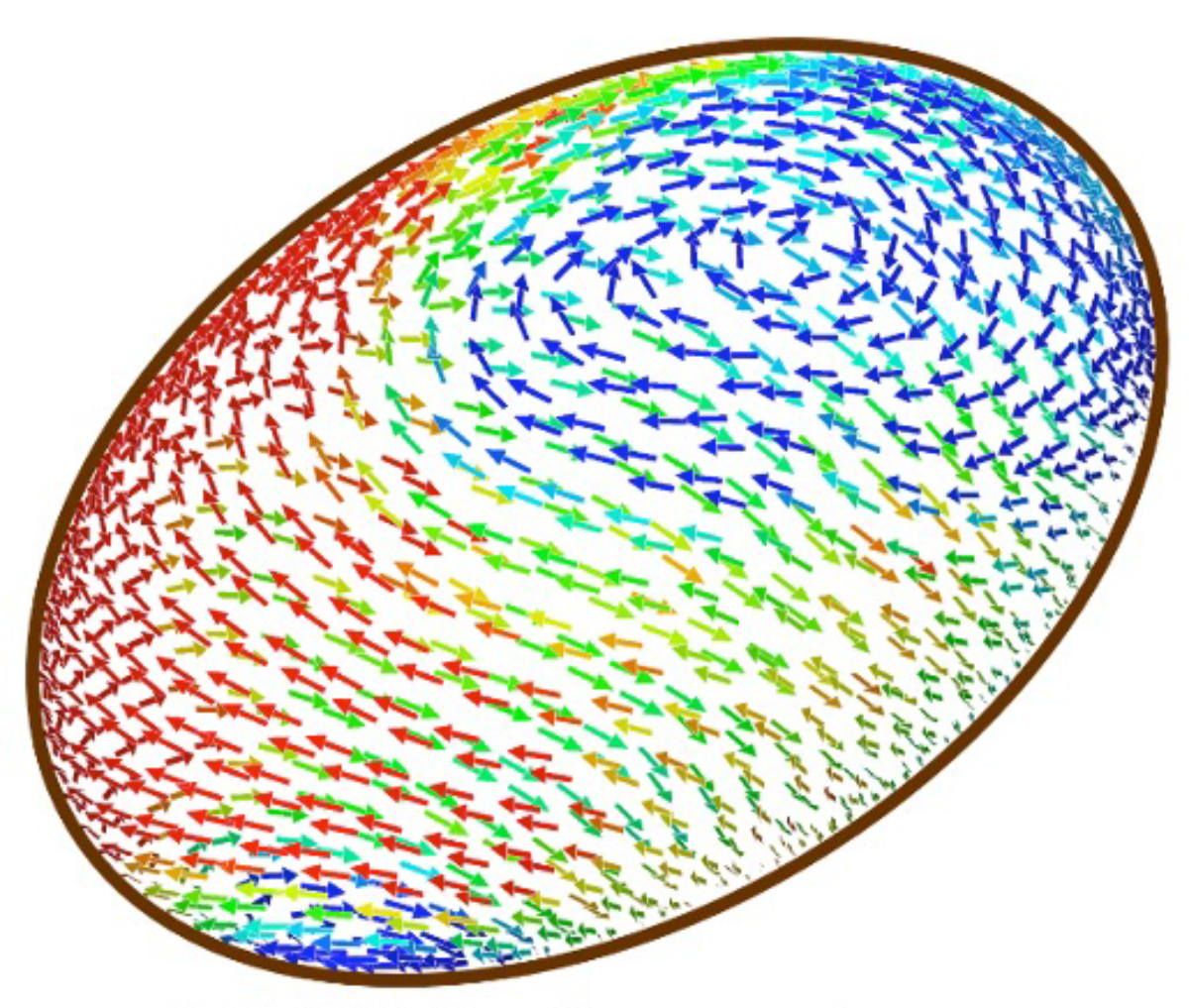
Figure: A snapshot of the simulation constructed in this study. The ellipsoid represents the fertilized egg of the nematode. The direction and speed of the flow near the surface of the cell are indicated by the direction and the colors of the arrows (red=fast, blue=slow). The image adopted from Fig. 2b of the paper.
Dam prevents the invasion of non-native freshwater shrimp into native habitats
Kitano Group / Ecological Genetics Laboratory
River dam prevents the invasion of non-native species of Neocaridina Kubo, 1938 (Decapoda: Caridea: Atyidae) into native habitats: A case study in the Yumesaki River system, Japan
Ryosuke Ishii and Yusuke Fuke *
* Corresponding author
Journal of Crustacean Biology (2025) 45, ruaf009 DOI:10.1093/jcbiol/ruaf009
The range expansion of invasive species that threaten biodiversity is caused by repeated cycles of their artificial introduction and subsequent dispersal. Dispersal can occur without human intervention, and its pattern is mainly determined by environmental factors and species characteristics.
In this study, Yusuke Fuke, JSPS postdoctoral fellow at the National Institute of Genetics, and Ryosuke Ishii, master’s student at Graduate School of Agriculture, Kagawa University, focused on the freshwater shrimp community in the Yumesaki River system, Japan, which is the habitat of the native species Neocaridina denticulata and has experienced the introduction of the closely related non-native species N. davidi. They examined the genetic population structures and morphological characteristics of these species to elucidate their dispersal process. Their results showed that populations comprising only the native species remained upstream of the dam, whereas the native species was replaced by the non-native species at the other sites. These results demonstrate that the distribution of non-native species can expand throughout the entire river system in rivers without structures that restrict the movement of aquatic organisms.

Figure: Native freshwater shrimp Neocaridina denticulata, found only upstream of the dam in the water system studied in this study.
A Near Complete Genome Assembly of the Oshima Cherry Cerasus speciosa
Press release
A Near Complete Genome Assembly of the Oshima Cherry Cerasus speciosa
Kazumichi Fujiwara, Atsushi Toyoda, Bhim B. Biswa, Takushi Kishida, Momi Tsuruta, Yasukazu Nakamura, Noriko Kimura, Shoko Kawamoto, Yutaka Sato, Toshio Katsuki, Sakura 100 Genome Consortium, and Tsuyoshi Koide*
Sakura 100 Genome Consortium (full list): Kazumichi Fujiwara, Atsushi Toyoda, Bhim B. Biswa, Takushi Kishida, Momi Tsuruta, Yasukazu Nakamura, Noriko Kimura, Shoko Kawamoto, Yutaka Sato, Toshio Katsuki, Tsuyoshi Koide*, Akatsuki Kimura, Ken-Ichi Nonomura, Hironori Niki, Hiroyuki Yano, Kinji Umehara, Tazro Ohta, Chikahiko Suzuki.
*Corresponding Author
Scientific Data volume 12, Article number: 162 (2025) DOI:10.1038/s41597-025-04388-z
![]() Press release (In Japanese only)
Press release (In Japanese only)
The Oshima cherry (Cerasus speciosa), which is endemic to Japan, has significant cultural and horticultural value. In this study, we present a near complete telomere-to-telomere genome assembly for C. speciosa, derived from the old growth “Sakurakkabu” tree on Izu Oshima Island. Using Illumina short-read, PacBio long-read, and Hi-C sequencing, we constructed a 269.3 Mbp genome assembly with a contig N50 of 32.0 Mbp. We examined the distribution of repetitive sequences in the assembled genome and identified regions that appeared to be centromeric. Detailed structural analysis of these putative centromeric regions revealed that the centromeric regions of C. speciosa comprised repetitive sequences with monomer lengths of 166 or 167 bp. Comparative genomic analysis with Prunus sensu lato genome revealed structural variations and conserved syntenic regions. This high-quality reference genome provides a crucial tool for studying the genetic diversity and evolutionary history of Cerasus species, facilitating advancements in horticultural research and the preservation of this iconic species.















