Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
“Metagenomic Thermometer” a novel approach to predict environmental temperatures based on metagenomic sequences.
Metagenomic Thermometer
Masaomi Kurokawa, Koichi Higashi, Keisuke Yoshida, Tomohiko Sato, Shigenori Maruyama, Hiroshi Mori, and Ken Kurokawa
DNA Research (2023) 30, dsad024 DOI:10.1093/dnares/dsad024
![]() Press release (In Japanese only)
Press release (In Japanese only)
Researchers at National Institute of Genetics have developed a groundbreaking tool, the “Metagenomic Thermometer,” which predicts environmental temperatures by analyzing the DNA of microorganisms in any given habitat. This innovative approach offers new insights into how environmental conditions influence life at a microbial level and could have wide-reaching implications in environmental science, biotechnology, and human health.
In a trailblazing study published in DNA Research, a team led by Professor Ken Kurokawa has unveiled the “Metagenomic Thermometer.” This tool marks a significant leap in environmental microbiology, enabling scientists to gauge the temperature of an environment by studying the genetic makeup of its microbial inhabitants.
The “Metagenomic Thermometer” operates on a groundbreaking principle: Professor Kurokawa’s team have extended the concept of estimating the optimal growth temperature (OGT) of a microorganism from the amino acid frequency of its genetic information, and developed a method of estimating the ambient temperature from the amino acid frequency in the collective genetic information of the microbial community present.
This revolutionary method was rigorously tested across diverse ecosystems, including hot springs, soil samples, and even the human gut. Remarkably, when applied to human gut metagenomic samples, the thermometer could accurately estimate human body temperature, underscoring the profound interplay between our internal environment and the microbial world.
Professor Kurokawa’s team believes that this tool doesn’t just measure temperature — it offers a new lens to understand how temperature drives the assembly of microbial communities. By focusing on amino acid composition rather than just microbial species, the Metagenomic Thermometer provides a more nuanced view of how life adapts to its environment.
The implications of this research are vast. From monitoring climate change impacts on microbial biodiversity to optimizing conditions for biotechnological applications, and even understanding human health in relation to our microbiome, the Metagenomic Thermometer stands as a testament to the power of innovative scientific inquiry.
“This research not only presents a novel tool but also opens up new avenues for understanding the intricate relationship between life and the environment,” said Professor Kurokawa. “We are excited about the potential applications of the Metagenomic Thermometer in various fields, including ecology, medicine, and biotechnology.”
- Variation in Gut Microbiome with Body Temperature:
The research proposes that body temperature significantly influences the gut microbiome composition. This revelation provides a deeper understanding of how bodily changes can impact gut health. - Predicting Deep Body Temperature:
Utilizing metagenomic analysis, the team has developed a ‘Metagenomic Thermometer’, which potentially allows for the estimation of human deep body temperature based on gut microbiota. This innovative approach offers a non-invasive method to gauge internal body temperature, a crucial health parameter. - Personalizing Medical Treatments:
The findings suggest exciting possibilities in tailoring treatments such as FMT and LBPs based on individual body temperatures. This could lead to more effective and personalized therapies for patients. - Probiotics Tailored to Body Temperature:
The research opens avenues for developing probiotics like yogurt and lactobacillus beverages specifically designed for individuals with varying body temperatures, enhancing their effectiveness. - Future Research Directions:
The team anticipates further exploration into how aging and diseases that lower body temperature affect gut microbiome composition. Additionally, the study’s methodologies could enhance predictions about bacterial community changes due to global warming, potentially influencing CO2 emission patterns.
For more information about this research, contact Office for Research Development.
– The study, “Metagenomic Thermometer,” was published in DNA Research.
– Interviews with the research team can be arranged upon request.
– High-resolution images and graphics explaining the Metagenomic Thermometer are available.
Contact:
Office for Research Development
National Institute of Genetics
National Institute of Genetics is a world-leading institution in genetics, genomics and bioinformatics. Our commitment to innovation and excellence places us at the forefront of global scientific endeavors.
Important notice regarding the entrance examination conducted in the winter of FY2023 (January 2024)
Taming the perils of photosynthesis – constraints on the evolution of the protective mechanisms
Miyagishima Group / Symbiosis and Cell Evolution Laboratory
Taming the perils of photosynthesis by eukaryotes: constraints on endosymbiotic evolution in aquatic ecosystems
Shin-ya Miyagishima
Communications Biology (2023) 6, 1150 DOI:10.1038/s42003-023-05544-0
An ancestral eukaryote acquired photosynthesis by genetically integrating a cyanobacterial endosymbiont as the chloroplast. The chloroplast was then further integrated into many other eukaryotic lineages through secondary endosymbiotic events of unicellular eukaryotic algae. While photosynthesis enables autotrophy, it also generates reactive oxygen species that can cause oxidative stress. To mitigate the stress, photosynthetic eukaryotes employ various mechanisms, including regulating chloroplast light absorption and repairing or removing damaged chloroplasts by sensing light and photosynthetic status. Recent studies have shown that, besides algae and plants with innate chloroplasts, several lineages of numerous unicellular eukaryotes engage in acquired phototrophy by hosting algal endosymbionts or by transiently utilizing chloroplasts sequestrated from algal prey in aquatic ecosystems. In addition, it has become evident that unicellular organisms engaged in acquired phototrophy, as well as those that feed on algae, have also developed mechanisms to cope with photosynthetic oxidative stress. These mechanisms are limited but similar to those employed by algae and plants. Thus, there appear to be constraints on the evolution of those mechanisms, which likely began by incorporating photosynthetic cells before the establishment of chloroplasts by extending preexisting mechanisms to cope with oxidative stress originating from mitochondrial respiration and acquiring new mechanisms.
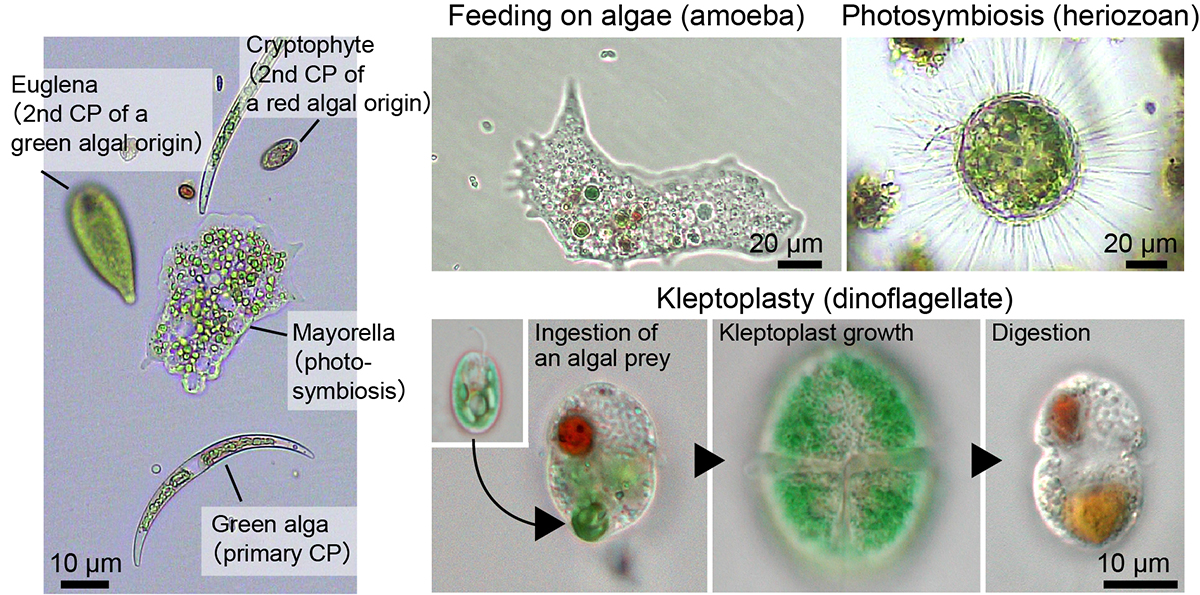
Figure: Examples of unicellular organisms with primary chloroplasts originating from cyanobacterial endosymbiosis, secondary chloroplasts from eukaryotic algal endosymbiosis, and organisms exhibiting phagotrophy, phototrophy, and kleptoplasty. Figure courtesy of Dr. Onuma (Kobe Univ.) and Mr. Okada (Nat. Inst. of Genet.).
A Practical Assembly Guideline for Genomes with Various Levels of Heterozygosity
Nakamura Group / Genome Informatics Laboratory
A Practical Assembly Guideline for Genomes with Various Levels of Heterozygosity
Takako Mochizuki, Mika Sakamoto, Yasuhiro Tanizawa, Takuro Nakayama, Goro Tanifuji, Ryoma Kamikawa, Yasukazu Nakamura*
*Corresponding Author
Briefings in Bioinformatics (2023) 24, bbad337 DOI:10.1093/bib/bbad337
The advancement of long-read sequencing technologies, exemplified by subreads of Pacific Biosciences, has significantly advanced our ability to reconstruct genome sequences. While these technologies can generate long reads, they are plagued by high sequence errors. To address these errors and strive to construct long, highly accurate contig sets, various de novo assemblers have been developed.
In de novo assembly of diploid genomes, the complexity increases with higher heterozygosity. Therefore, heterozygosity is a significant factor influencing the completeness of de novo assembly. However, systematic evaluations of de novo assemblers for diploid genomes with various heterozygosity levels have not been conducted.
In this study, using genomes with varying levels of heterozygosity, we conducted a series of processes, including estimation of genome characteristics such as genome size and heterozygosity, de novo assembly, polishing, and removing contigs including alleles. We have presented a guideline for constructing a representative haplotype set based on heterozygosity levels.
This work was supported by JSPS grant-in-aid for Scientific Research on Innovative Areas, Platform for Advanced Genome Science [16H06279], and KAKENHI [15H05606 and 19H03274, 20H03305, 17H03723].
Computations were partially performed on the National Institute of Genetics supercomputer.
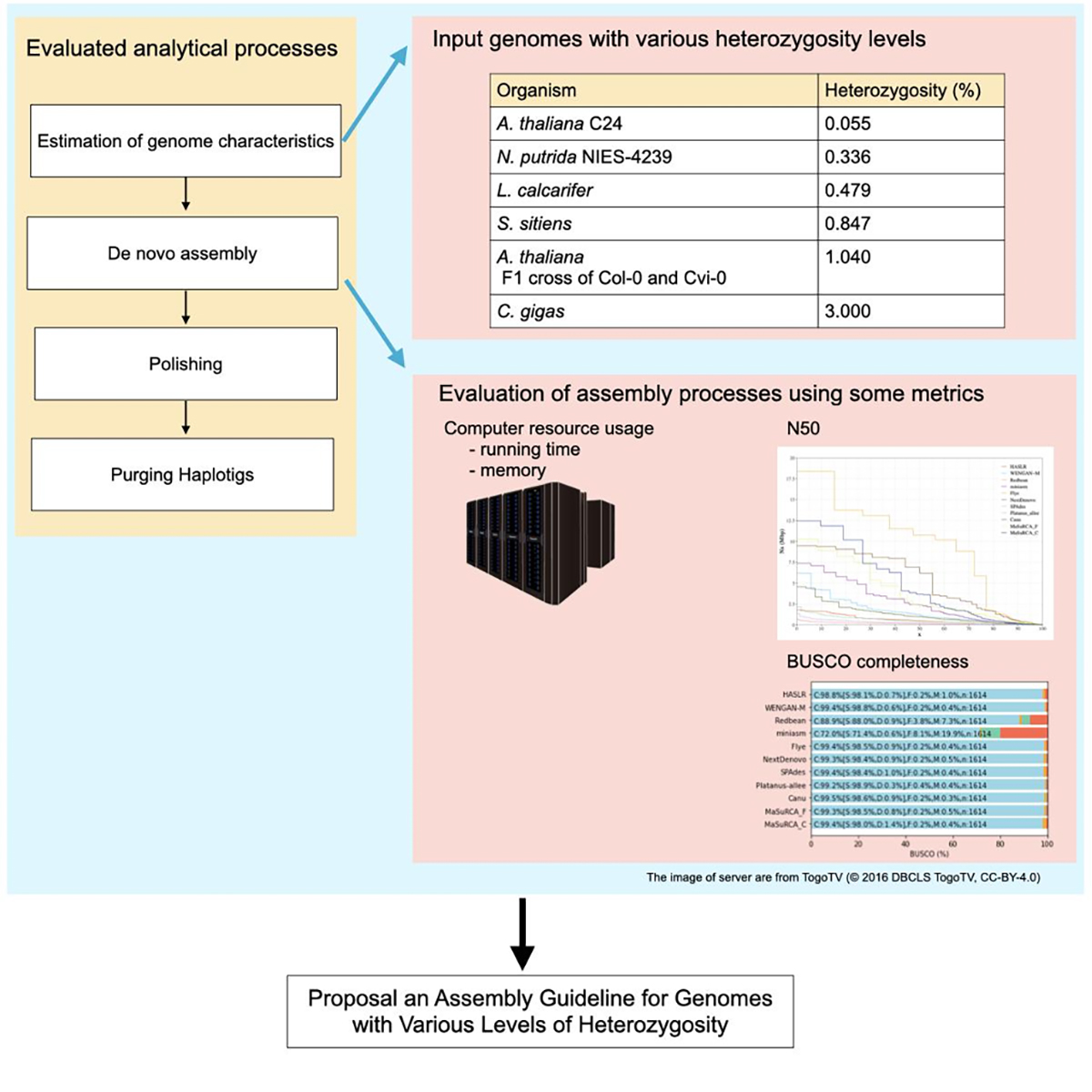
Figure: An evaluation process for genome assembly using genomes with various levels of heterozygosity
First Japanese record of freshwater prawn from Ryukyu Archipelago
Kitano Group / Ecological Genetics Laboratory
First record of Macrobrachium mammillodactylus (Thallwitz, 1891) (Crustacea, Decapoda, Palaemonidae) from Japan.
Yusuke Fuke, Tomoaki Maruyama.
Check List (2023) 19, 821-826 DOI:10.15560/19.6.821
The freshwater prawn genus Macrobrachium has more than 270 known species, with remarkably high species diversity in tropical freshwaters. Most of the widely distributed species in the Pacific have amphidromous migratory life history, which allows them to expand their range via ocean currents.
In this study, Yusuke Fuke, JSPS Postdoctoral Fellow at the National Institute of Genetics, and Tomoaki Maruyama of Trend Design Co., Ltd. identified the specimens collected from Miyako Island as Macrobrachium mammillodactylus (Thallwitz, 1891) based on genetic and morphological analysis. This marks the first record of this species in Japan and its northernmost distribution. This species has been previously reported from Australia to central Taiwan. The specimens collected in this study were adults, suggesting that they likely overwintered on the island, indicating an expansion of their range to the Ryukyu Archipelago.
Range expansion from Southeast Asia to the Ryukyu Archipelago and Honshu has been observed in other southern freshwater shrimp species. By accumulating fundamental range data, we may be able to identify and predict the factors contributing to the range expansion of amphidromous species.

Figure: Macrobrachium mammillodactylus collected from Miyako Island, Japan. Photo by T. Maruyama.















