Condensins and nucleosome–nucleosome interactions differentially constrain chromatin to organize mitotic chromosomes
Press release
Single-nucleosome imaging unveils that condensins and nucleosome-nucleosome interactions differentially constrain chromatin to organize mitotic chromosomes.
Kayo Hibino, Yuji Sakai, Sachiko Tamura, Masatoshi Takagi, Katsuhiko Minami, Masa A. Shimazoe, Toyoaki Natsume, Masato T. Kanemaki, Naoko Imamoto, Kazuhiro Maeshima*
* Corresponding Author
Nature Communications (2024) 15, 7152 DOI:10.1038/s41467-024-51454-y
![]() Press release (In Japanese only)
Press release (In Japanese only)
For accurate mitotic cell division, replicated chromatin must be assembled into chromosomes and faithfully segregated into daughter cells. While protein factors like condensin play key roles in this process, it is unclear how chromosome assembly proceeds as molecular events of nucleosomes in living cells and how condensins act on nucleosomes to organize chromosomes. To approach these questions, we investigate nucleosome behavior during mitosis of living human cells using single-nucleosome tracking, combined with rapid-protein depletion technology and computational modeling. Our results show that local nucleosome motion becomes increasingly constrained during mitotic chromosome assembly, which is functionally distinct from condensed apoptotic chromatin. Condensins act as molecular crosslinkers, locally constraining nucleosomes to organize chromosomes. Additionally, nucleosome-nucleosome interactions via histone tails constrain and compact whole chromosomes. Our findings elucidate the physical nature of the chromosome assembly process during mitosis.
This study was published online as open access in Nature Communications on August 21, 2024.
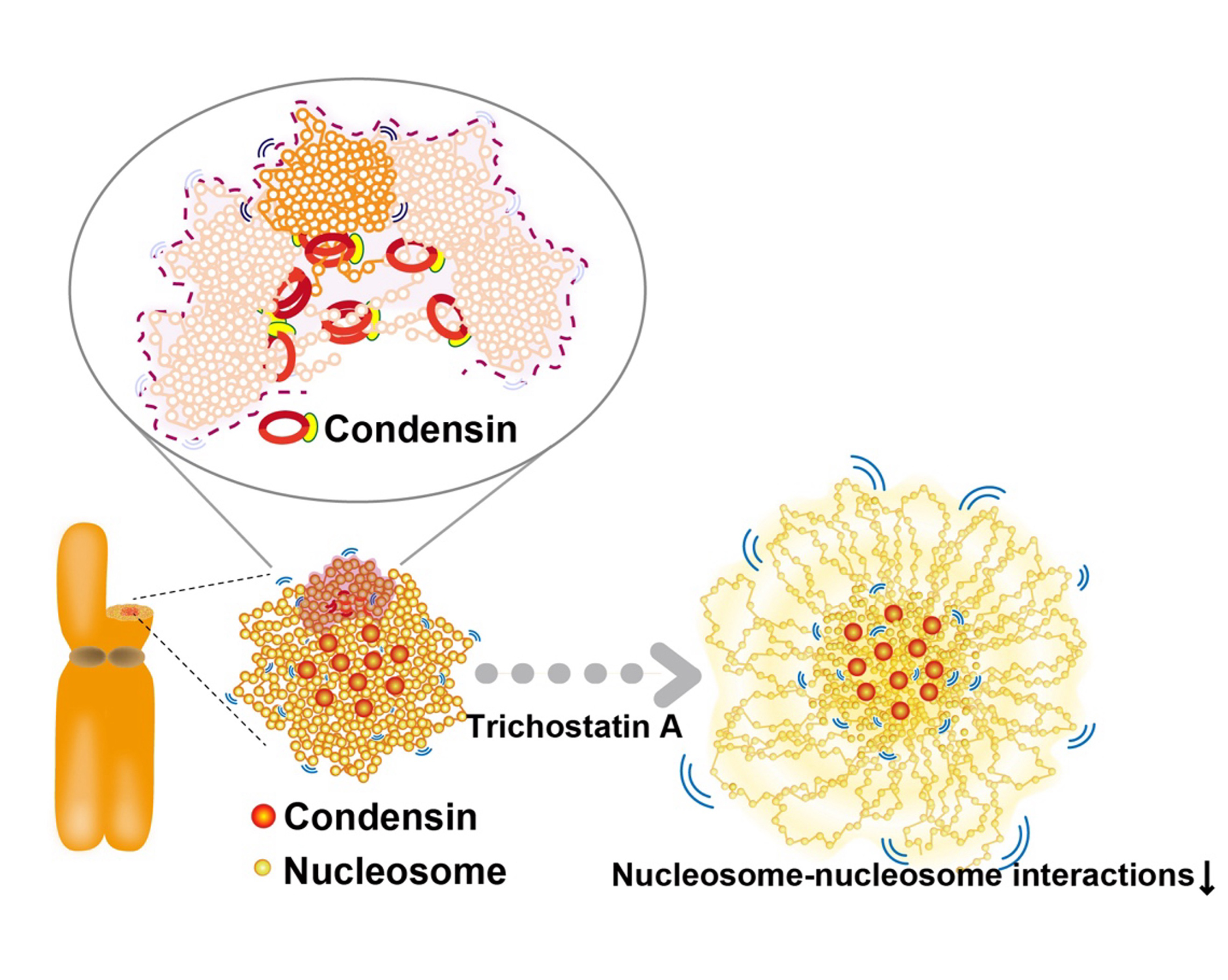
Figure: Condensins seem to act as a molecular crosslinkersto make loops. (Bottom, left) Condensins (red) locate around chromosome center. (Bottom, right) Nucleosomes around the periphery (those mostly free from condensins) in the Trichostatin A-treated chromosomes, whose nucleosome–nucleosomes interactions are weakened, are less constrained and have higher mobility than those around the axis.
Video: Single nucleosomes fluorescently labeled in a living HeLa interphase cell (left) and mitotic cell (center)(50 ms/frame). Each dot represents a single nucleosome. (Right) Calculated chromatin motion of the simulated chromosome. Chromatin beads (blue) and condensins (green and red) are shown.















