Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
The dwarf neon rainbowfish Melanotaenia praecox, a small spiny-rayed fish with potential as a new Acanthomorpha model fish: II. Establishment of a microinjection procedure for genetic engineering
Kawakami Group / Laboratory of Molecular and Developmental Biology
The dwarf neon rainbowfish Melanotaenia praecox, a smallspiny-rayed fish with potential as a new Acanthomorphamodel fish: II. Establishment of a microinjection procedurefor genetic engineering.
Kazuhide Miyamoto, Gembu Abe, Koichi Kawakami, Koji Tamura, Satoshi Ansai
Developmental Dynamics 2024 Feb 5 DOI:10.1002/dvdy.698
Background: Rainbowfish is a clade of colorful freshwater fish. Melanotaenia praecox is a small rainbowfish species with biological characteristics that make it potentially useful as an experimental model species. We anticipate that M. praecox could become a new model used in various fields, such as ecology, evolution, and developmental biology. However, few previous studies have described experimental set-ups needed to understand the molecular and genetic mechanisms within this species. Results: We describe detailed procedures for genetic engineering in the rainbowfish M. praecox. By using these procedures, we successfully demonstrated CRISPR/Cas-mediated knockout and Tol2 transposon-mediated transgenesis in this species. Regarding the CRISPR/Cas system, we disrupted the tyrosinase gene and then showed that injected embryos lacked pigmentation over much of their body. We also demonstrated that a Tol2 construct, including a GFP gene driven by a ubiquitous promoter, was efficiently integrated into the genome of M. praecox embryos. Conclusions: The establishment of procedures for genetic engineering in M. praecox enables investigation of the genetic mechanisms behind a broad range of biological phenomena in this species. Thus, we suggest that M. praecox can be used as a new model species in various experimental biology fields.
This study was conducted as collaboration with Tamura lab at Tohoku University.
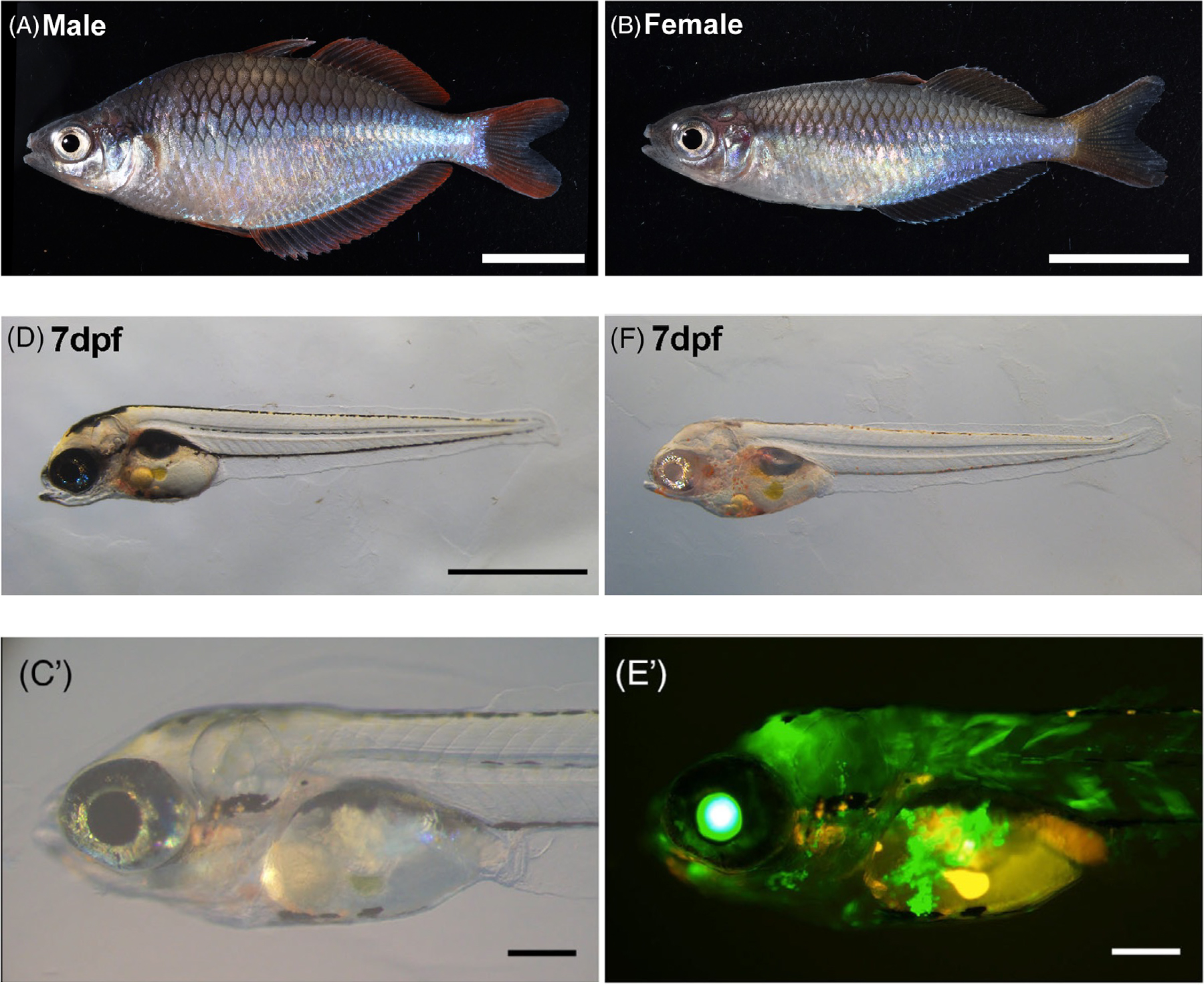
Figure: (A)(B) Male and female Melanotaenia praecox.
(D)(F) Disruption of the tyrosinase gene using CRISPR/Cas9.
(C’)(E’) Introduction of the GFP gene using Tol2.
More constrained chromatin in oncogenic HRAS-transformed cells.
Maeshima Group / Genome Dynamics Laboratory
Kurokawa Group / Genome Evolution Laboratory
Kuraku Group / Molecular Life History Laboratory
Chromatin organization and behavior in HRAS-transformed mouse fibroblasts
Aoi Otsuka*, Katsuhiko Minami*, Koichi Higashi, Akane Kawaguchi, Sachiko Tamura, Satoru Ide, Michael J. Hendzel, Ken Kurokawa, Kazuhiro Maeshima#
* These authors equally contributed to this work. #corresponding author
Chromosoma 2024 Feb 24 DOI:10.1007/s00412-024-00817-x
Genomic DNA is organized three-dimensionally in the nucleus as chromatin. Recent super-resolution imaging and Hi-C studies have shown that chromatin in living cells forms a number of condensed chromatin domains. Inside these domains, nucleosomes behave locally like a liquid, and such behavior is deeply related to various DNA functions. Cancer cells often show upregulated transcription and other activities, but how is the chromatin behavior in cancer cells different from non-cancer cells?
A research team led by Professor Kazuhiro Maeshima of Genome Dynamics Laboratory (NIG), including a SOKENDAI graduate student Aoi Otsuka (SOKENDAI Special Researcher), SOKENDAI graduate student Katsuhiko Minami (former SOKENDAI Special Researcher, JSPS Research Fellow DC2), Assistant Professor Satoru Ide, Technical Staff Sachiko Tamura, together with Assistant Professor Koichi Higashi and Professor Ken Kurokawa of Genome Evolution Laboratory (NIG), Assistant Professor Akane Kawaguchi of Molecular Life History Laboratory (NIG), and Professor Michael J. Hendzel of University of Alberta, have found chromatin in oncogenic RAS-transformed cells is more constrained than their parental cells.
In this work, the research team examined the chromatin behavior in mouse oncogenic HRAS-transformed cells (CIRAS-3 cells) and their parental cells (10T1/2 cells) with multiple approaches. First, they found that the CIRAS-3 cells have smaller nuclei. In addition, they investigated the individual nucleosome movements in living cells using super-resolution fluorescence microscopy and revealed that nucleosomes are locally more constrained in CIRAS-3 cells. Consistently, CIRAS-3 cells show increased heterochromatin, and in situ Hi-C revealed enriched interactions of the B-B compartments, which mainly consist of heterochromatin.
Genome chromatin in cancer cells encounters physical stress during migration through small spaces, such as metastasis. The smaller nuclei and the more constrained chromatin with enriched heterochromatin can gain elastic properties of chromatin, and may better benefit the cell metastasis (Figure).
This work was supported by JSPS Fellowship, JSPD grants (21H02453, 23K17398, 23K05798, 22H05606, 21H02535, 23KJ0998, 20H05936, 22H04925 (PAGS)), a Japan Science and Technology Agency JST SPRING(JPMJSP2104), The Naito Foundation, and the Takeda Science Foundation. Genome analysis was performed with support of Platform for Advanced Genome Science (22H04925 (PAGS)).
The journal “Chromosoma” is a historical chromosome journal first published in 1939, and is available from the first issue in the NIG Library.
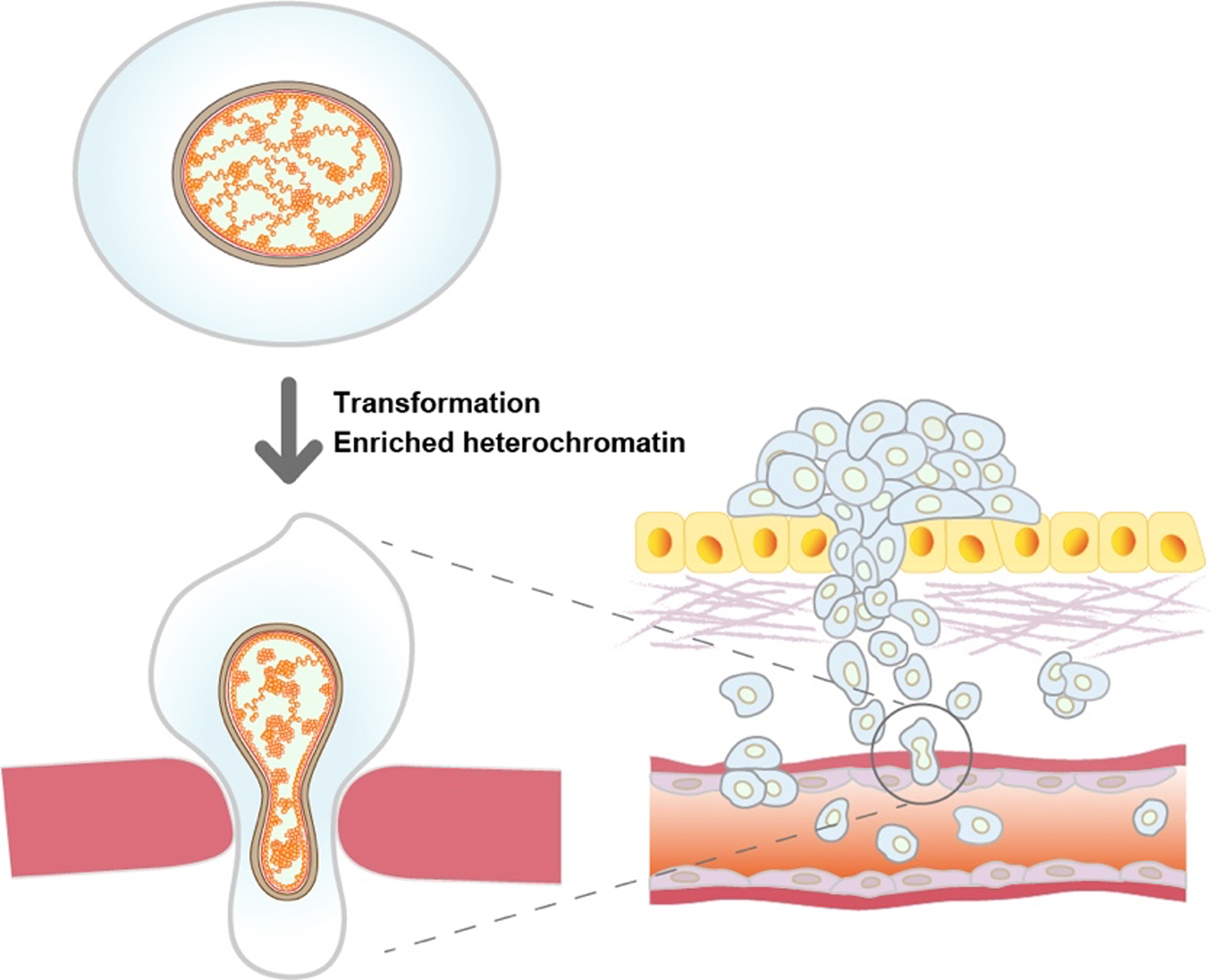
Figure: Oncogenic HRAS-transformed cells (bottom left) have more constrained chromatin with increased heterochromatin than non-transformed parental cells (top left). The more constrained chromatin may play an important role in the metastasis process.
Association of tameness and sociability but no sign of domestication syndrome in mice selectively bred for active tameness
KoideGroup / Mouse Genomics Resource Laboratory
Association of tameness and sociability but no sign of domestication syndrome in mice selectively bred for active tameness
Bharathi Venkatachalam, Bhim B. Biswa, Hiromichi Nagayama, Tsuyoshi Koide*
*責任著者
Genes, Brain and Behavior (2024) 23, e12887 DOI:10.1111/gbb.12887
Domesticated animals have been developed by selecting desirable traits following the initial unconscious selection stage, and now exhibit phenotypes desired by humans. Tameness is a common behavioural trait found in all domesticated animals. At the same time, these domesticated animals exhibit a variety of morphological, behavioural, and physiological traits that differ from their wild counterparts of their ancestral species. These traits are collectively referred to as domestication syndrome. However, whether this phenomenon exists is debatable. Previously, selective breeding has been used to enhance active tameness, a motivation to interact with humans, in wild heterogeneous stock mice derived from eight wild inbred strains. In the current study, we used tame mice to study how selective breeding for active tameness affects behavioural and morphological traits. A series of behavioural and morphological analyses on mice showed an increased preference for social stimuli and a longer duration of engagement in non-aggressive behaviour. However, no differences were observed in exploratory or anxiety-related behaviours. Similarly, selection for tameness did not affect ultrasonic vocalisations in mice, and no changes were observed in known morphological traits associated with domestication syndrome. These results suggest that there may be a link between active tameness and sociability and provide insights into the relationship between tameness and other behaviours in the context of domestication.
Source: Bharathi Venkatachalam et al., Genes, Brain and Behavior (2024) 23, e12887
Ensuring personal space inside the cell
Kimura Group / Cell Architecture Laboratory
Enucleation of the C. elegans embryo revealed dynein-dependent spacing between microtubule asters
Fujii, K., Kondo, T. & *Kimura
*corresponding author
Life Sci. Alliance (2023) 7, e202302427 DOI:10.26508/lsa.202302427
In psychology, personal space is known as the distance that people tend to maintain from others. A similar phenomenon can be found inside the cell. An organelle called centrosome tend to maintain certain distance from the other centrosomes. In this study, Dr. Ken Fujii and his colleagues investigated the behavior of the centrosomes in enucleated, C. elegans embryonic cells. Using genetics and computational approaches, the researchers proposed that the centrosomes compete with each other for motor proteins “dynein” distributed in the cytoplasm and at the cell cortex to maintain certain distance from the others and ensure their “personal space.” The study has implication in how cellular structures measure distances inside the cell.
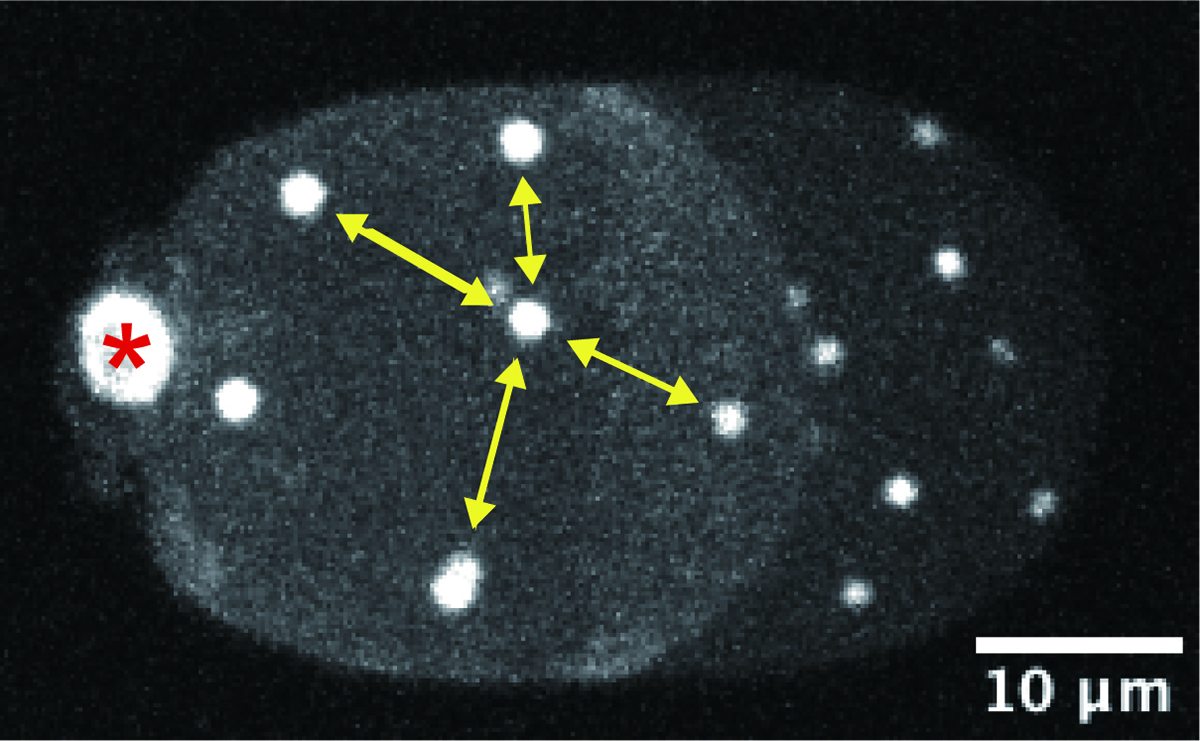
Figure: A representative microscope image of an enucleated, C. elegans embryonic cell. White signals indicate the centrosomes, which are evenly distributed inside the cell. (The image is a 2-dimensional projection of the 3-dimensional cell image. The asterisk indicates the polar body, which is not the centrosome.)
Target-selective vertebrate motor axon regeneration depends on interaction with glial cells at a peripheral nerve plexus
Kawakami Group / Laboratory of Molecular and Developmental Biology
Target-selective vertebrate motor axon regeneration depends on interaction with glial cells at a peripheral nerve plexus
Lauren J. Walker, Camilo Guevara, Koichi Kawakami, and Michael Granato
PLOS Biology (2023) 21, e3002223 DOI:10.1371/journal.pbio.3002223
A critical step for functional recovery from peripheral nerve injury is for regenerating axons to connect with their pre-injury targets. Reestablishing pre-injury target specificity is particularly challenging for limb-innervating axons as they encounter a plexus, a net-work where peripheral nerves converge, axons from different nerves intermingle, and then resort into target-specific bundles. Here, we examine this process at a plexus located at the base of the zebrafish pectoral fin, equivalent to tetrapod forelimbs. Using live cell imaging and sparse axon labeling, we find that regenerating motor axons from 3 nerves coalesce into the plexus. There, they intermingle and sort into distinct branches, and then navigate to their original muscle domains with high fidelity that restores functionality. We demonstrate that this regeneration process includes selective retraction of mistargeted axons, suggesting active correction mechanisms. Moreover, we find that Schwann cells are enriched and associate with axons at the plexus, and that Schwann cell ablation during regeneration causes profound axonal mistargeting. Our data provide the first real-time account of regenerating vertebrate motor axons navigating a nerve plexus and reveal a previously unappreciated role for Schwann cells to promote axon sorting at a plexus during regeneration.
This study was conducted as collaboration with Granato lab at University of Pennsylvania.
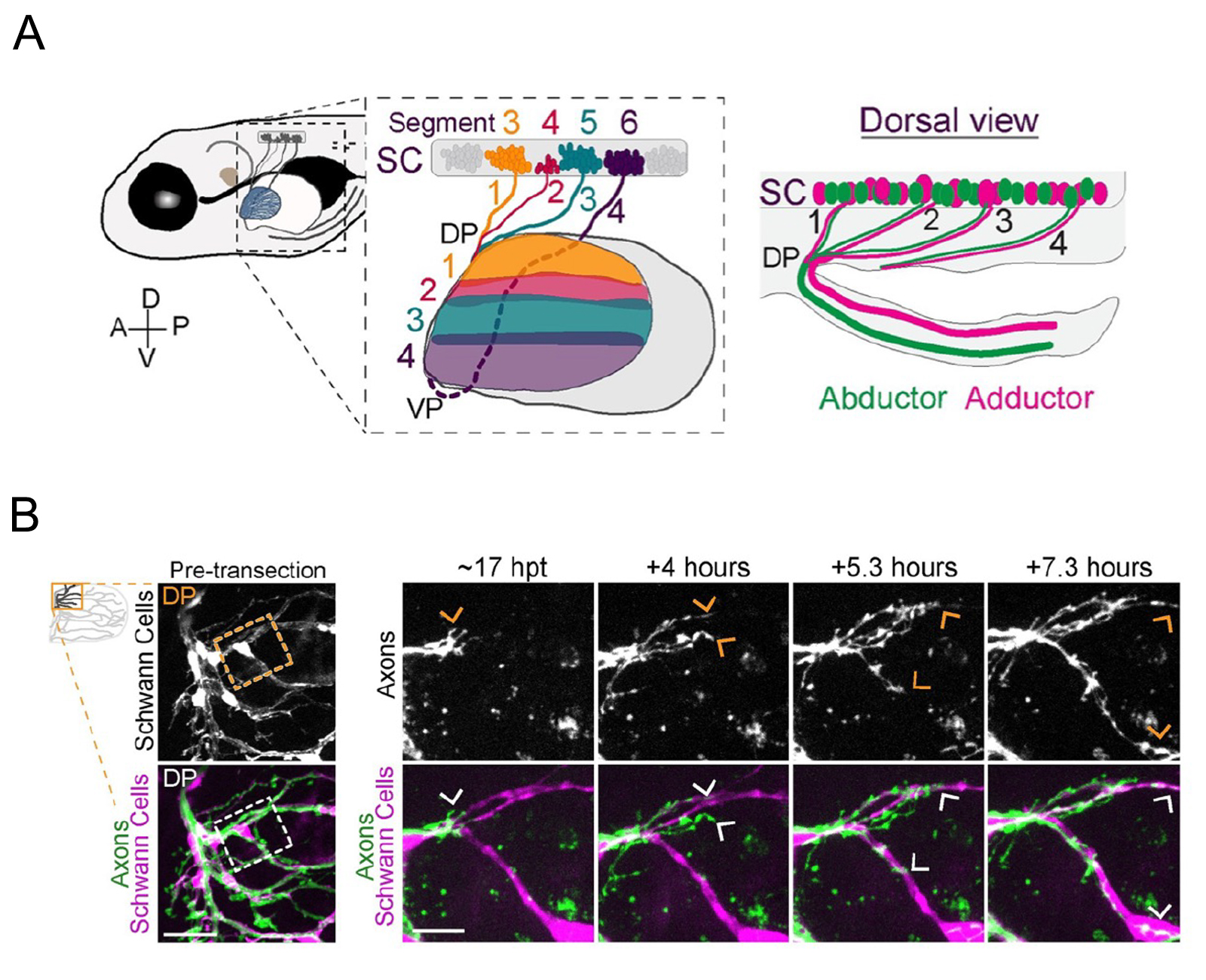
Figure: (A)Zebrafish spinal motor nerves are resorted in a plexus (DP and VP ) and connect to the muscle of the pectoral fin.
(B)Live imaging of navigation of axons along Schwann cells. Transgenic zebrafish were used to label Schwann cells.















