Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
The Start of Whole Genome Analysis of Novel Coronavirus Based on the Cooperation Request from Shizuoka Prefecture
Infection of novel coronavirus (SARS-CoV-2) variants is spreading mainly in large cities. In response to the spread of novel coronavirus (COVID-19), we, National Institute of Genetics, have offered cooperation to Shizuoka prefecture in April 2020 and have concluded an agreement on investigation cooperation in July.
We will start analyzing genome information (whole genetic information) of the specimens collected from COVID-19 patients in the prefecture as early as this month upon further cooperation request from Shizuoka prefecture. Specimen samples (nucleic acid substance) provided by the prefecture are not infectious and safe. We quickly determine presence/absence of variant virus by analyzing the genome information of the samples and report the result to the prefecture.
Genome sequencing of those samples enables us to accurately detect and identify SARS-CoV-2 variants spreading within the prefecture and to contributes to the prevention measures against COVID-19 by using genome sequence information.
SARS-CoV-2 genome sequence decoded by this research investigation will be released through the INSD (the International Nucleotide Sequence Database) and GISAID (Global Initiative on Sharing Avian Influenza Data). In the flamework of open science, our institute will actively cooperate with other projects on anti-virus measures conducted by the government.
April 26th, 2021
HANAOKA, Fumio
Director-General, National Institute of Genetics
60-year scientific mystery of ’the DNA replication-timing program’ has finally been solved!
Replication timing maintains the global epigenetic state in human cells
Kyle N Klein, Peiyao A Zhao, Xiaowen Lyu, Takayo Sasaki, Daniel A Bartlett, Amar M Singh, Ipek Tasan, Meng Zhang, Lotte P Watts, Shin-Ichiro Hiraga, Toyoaki Natsume, Xuemeng Zhou, Timour Baslan, Danny Leung, Masato T Kanemaki, Anne D Donaldson, Huimin Zhao, Stephen Dalton, Victor G Corces, David M Gilbert
Science 372, 371-378 (2021) DOI:10.1126/science.aba5545
The temporal order of DNA replication is conserved from yeast to humans, but its biological significance remains unclear. We eliminated the protein RIF1, a master regulator of replication timing, in several human cell lines. RIF1 loss during the G1 phase of the cell cycle resulted in a heterogeneous, nearly random replication timing program from the first S phase. Altered replication timing was followed by replication-dependent redistribution of active and repressive histone modifications and alterations in genome architecture. These results support a model in which replication timing orchestrates the epigenetic state of newly replicated chromatin.
The Kanemaki laboratory contributed to the generation of a conditional RIF1 mutant using the auxin-inducible degron system.
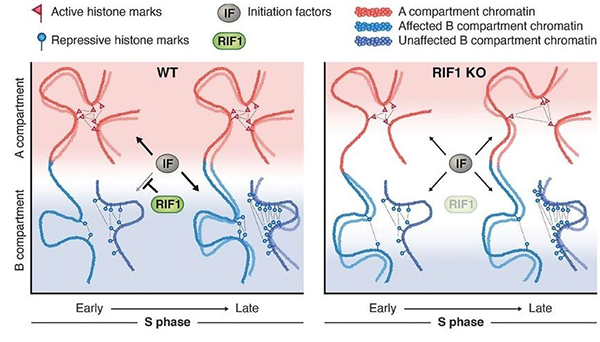
Figure: A model showing the role of the replication-timing program for the maintenance of the epigenome.
- You can read the “EurekAlert!” article here.
Prof. Maeshima’s comment was published in Molecular Cell. “New door to understanding transcription, open with caution.”
 Prof. Maeshima’s comment on the role of phase separation in transcription, “New door to understanding transcription, open with caution.” was published in the April 15 issue of Molecular Cell (Cell Press).
Prof. Maeshima’s comment on the role of phase separation in transcription, “New door to understanding transcription, open with caution.” was published in the April 15 issue of Molecular Cell (Cell Press).
![]() https://www.cell.com/molecular-cell/fulltext/S1097-2765(21)00267-7
https://www.cell.com/molecular-cell/fulltext/S1097-2765(21)00267-7
In recent years, a principle called “liquid-liquid phase separation (LLPS)” has been in the limelight in cell biology. It is thought that LLPS can increase the concentration of specific molecules in cells and create membrane-less structures “droplets”, making it possible to control cellular functions spatially and temporally. Because of this trend, many researchers claim that the intracellular assembly of various molecules is a droplet.
This time, while Prof. Maeshima admits that LLPS is an exciting new concept in cell biology, he argues that there is a lack of tools to prove the existence of “droplets” in the cell and that careful research based on the observation of individual molecule behavior is required.
Mechanisms to stabilize genome by chromatin remodeling
Kakutani Group / Epigenomics Laboratory
The chromatin remodeler DDM1 prevents transposon mobility through deposition of histone variant H2A.W
Akihisa Osakabe, Bhagyshree Jamge, Elin Axelsson, Sean A. Montgomery, Svetlana Akimcheva, Annika Luisa Kuehn, Rahul Pisupati, Zdravko J. Lorković, Ramesh Yelagandula, Tetsuji Kakutani, and Frédéric Berger
Nature Cell Biology 23, 391-400 (2021) DOI:10.1038/s41556-021-00658-1
Transposable elements (TEs) are generally silenced by epigenetic modifications such as methylation of DNA and histone. An Arabidopsis chromatin remodeler protein DDM1 (Decrease in DNA methylation 1) was identified by genetic screening as a factor necessary for maintenance of these silencing marks. However, the underlying mechanisms of this function of DDM1 remain unknown.
In this study, we focused on histone variant, H2A.W, which localizes at TE-rich heterochromatic regions. Our genomic analysis revealed that H2A.W was lost from heterochromatin when DDM1 is non-functional. DDM1 directly binds to H2A.W. Histone binding regions of DDM1 are important for H2A.W deposition and TE silencing. These results suggest that DDM1 silences TEs by deposition of H2A.W into heterochromatin. This pathway to silence TEs by chromatin remodeling could be conserved to mammals, since mammals have DDM1 and H2A.W orthologs with similar functions.
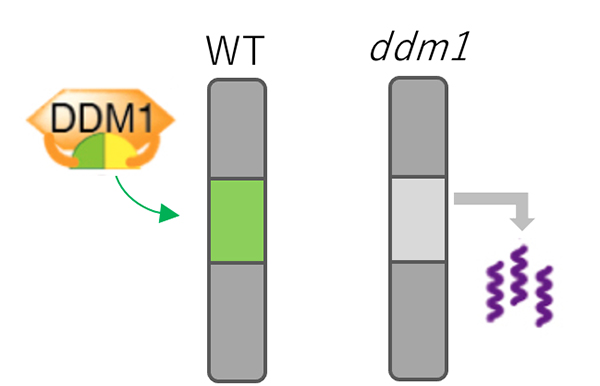
Figure: Mechanism for TE silencing regulated by DDM1.
In wild type plant (left), DDM1 mediates deposition of H2A.W (shown by green) into genomic regions with silent TEs. In ddm1 mutants (right), H2A.W was lost from heterochromatin, which is associated with derepression of TEs.
Six papers published on March 31, 2021 in “Anthropological Science” special issue on Yaponesian Genome Project
Press release
0 (preface).
Preface to the special issue on the Yaponesia Genome Project
Naruya Saitou
1 (original article).
Modern human DNA analyses with special reference to the Inner-Dual structure model of Yaponesian
Timothy Jinam, Yosuke Kawai, and Naruya Saitou
2 (original article).
Ancient genomes from the initial Jomon period: new insights into the genetic history of the Japanese archipelago
Noboru Adachi, Hideaki Kanzawa-Kiriyama, Nara T., Kakuda T., Nishida I., and Ken-ichi Shinoda
3 (original article).
The time-dependent evolutionary rate of mitochondrial DNA in small mammals inferred from biogeographic calibration points with reference to the late Quaternary environmental changes
Hitoshi Suzuki
4 (original article).
Geographical distribution of certain toponyms in the Samguk Sagi
Mitsuaki Endo
5 (review article).
Exploring models of human migration to the Japanese archipelago using genome-wide genetic data
Naoki Osada and Yosuke Kawai
6 (review article).
Paleogenomics of human remains in East Asia and Yaponesia focusing on current advances and future directions
Kae Koganebuchi and Hiroki Oota
Abstract of Paper 1:
Previous studies suggested two major migration events during the Jomon and Yayoi periods that affected the genetic diversity of modern Japanese (Yaponesians). We explored the possibility of a three-wave migration model by examining three datasets of modern human DNA: (1) whole mitochondrial (mt) DNA genomes of 1642 Yaponesians; (2) mtDNA haplogroup frequencies of 59105 Yaponesians from 47 prefectures; and (3) genome-wide SNP data of two Yaponesians (Ainu, Okinawa) and whole-genome sequence data of Yamato individuals, the Funadomari Jomon F23 individual, and three East Asian populations (Korean, northern Chinese, and southern Chinese). Past population size change was estimated based on dataset 1, and we clearly observed a steep population increase after the Yayoi period. Principal-component analysis and phylogenetic network analysis were applied to dataset 2, and we confirmed the pattern consistent with our model. An admixture program was used on dataset 3, and we found that the two- and three-layer migration models are both compatible with these SNP data. Taken together, these three datasets provide support for our three-wave, ‘inner dual-structure’ model.
Abstract of Paper 2:
Starting 16000 years ago, the Neolithic lifestyle known as the Jomon culture spread across the Japanese archipelago. Although extensively studied by archaeology and physical anthropology, little is known about the genetic characteristics of the Jomon people. Here, we report the entire mitogenome and partial nuclear genome of skeletal remains from the initial Jomon period that were excavated from the Higashimyo shell midden site at Saga City, Kyushu Island, Japan. This is the first genome analysis of the initial Jomon people of Kyushu Island. These results provide important data for understanding the temporal transition and regional differences of the Jomon people. The mitochondrial DNA and Y-chromosome haplogroups were similar to those found in the previously reported later Jomon people. Moreover, comparison of three nuclear genomes from the initial to final Jomon periods indicated genetic continuity throughout the Jomon period within the Japanese archipelago with no significant evidence of admixture. This indicates that the genetic differentiation found among the Jomon people was promoted by the progression of regionalization throughout the Jomon period. Further accumulation of high-quality Jomon genome data spanning a wide range of regions and ages will clarify both intimate regional and temporal differences of the Jomon people and details of their admixture history with rice farmers, as suggested by Jomon mitochondrial genome data. The results obtained from this study provide important information for further analysis.
Abstract of Paper 3:
Mitochondrial DNA (mtDNA) sequences have long been the most popular marker for assessing phylogenetic relationships and uncovering population dynamics. However, the mechanism of the nucleotide substitution rate of mtDNA remains unclear. While the evolutionary rate over tens of thousands of years is thought to be time dependent, the overall picture is not fully understood. This article presents recent achievements related to the time-dependent evolutionary rate of mtDNA in small rodents in the Japanese archipelago. The method focuses on rapid expansion events during the late Quaternary, during which there was a prolonged severe cold period and repeated abrupt warm periods, providing multiple calibration points. The global sea level fluctuation and migration to islands help to specify the calibration points. For calibration points at 11000, 15000, 53000, and 130000 years ago, the evolutionary rates were approximately 0.11, 0.11, 0.047, and 0.029 substitutions/site/million years, respectively, in the mitochondrial cytochrome b gene (Cytb). Applying the higher rate to assess the evolutionary history of the commensal house mouse (Mus musculus) and complete mitochondrial genome sequences (~16000 bp) allowed us to trace prehistoric human culture development based on millet and rice agriculture. The pattern of time-dependent evolutionary rates presented here is likely applicable to other small rodents. The Japanese archipelago is ideal for assessing evolutionary rates with biogeographic calibration points in the late Quaternary in species with multiple genetically distinct local populations.
Abstract of Paper 4:
Kim Busik’s Samguk Sagi 三国史記, History of the Three Kingdoms, dating from 1145 A.D., is renowned for including Japanese toponyms in the Korean peninsula and the north of Yalujiang district (modern Liaoning and Jilin Provinces). Kim’s work recorded the older, archaic toponyms before they were converted into sinicized (i.e. expressed with Chinese words) place names with two Chinese characters in 757 A.D. by the order of King Kyeongdeok. This paper maps specific words included in the place names of 783 locations for which the corresponding present places are evident. The following were words examined: (1) ‘river’ and its related words; (2) ‘valley’; (3) ‘mountain’ and ‘ridge’; and (4) ‘city’ and ‘burg’. Japonic-sourced toponyms are typically distributed in the central and northern areas of the Yalu River, primarily in the district of Koguryŏ; however, they go beyond these regions. The use of Chinese loanwords is noted in the southern area, where determining which language was spoken is difficult. In a town near Seoul, the stem of the toponym belongs to the Korean language, whereas the unit word belongs to the Japonic language. This usage may be attributed to bilingualism, whereby Korean-speaking inhabitants used their own language for the stem of the place name. Mongolic and/or Tungusic loanwords are also found. In some cases, determining the language origin of the current toponyms is difficult. Therefore, the minute geographical distribution of the origin languages is displayed word by word. These toponyms reflect the traces of indigenous languages and reveal that Japonic-speaking people still dwelled in the central area of the peninsula and in the northern area of the Yalu River at that period.
Abstract of Paper 5:
The origins of people in the Japanese archipelago are of long-standing interest among anthropologists, archeologists, linguists, and historians studying the history of Japan. While the ‘dual-structure’ model proposed by Hanihara in 1991 has been considered the primary working hypothesis for three decades, recent advances in DNA typing and sequencing technologies provide an unprecedented amount of present-day and ancient human nuclear genome data, which enable us to refine or extend the dual-structure model. In this review, we summarize recent genome sequencing efforts of present-day and ancient people in Asia, mostly focusing on East Asia, and we discuss the possible migration routes and admixture patterns of Japanese ancestors. We also report on a meta-analysis we performed by compiling publicly available datasets to clarify the genetic relationships of present-day and ancient Japanese populations with surrounding populations. Because the ancient genetic data from the Japanese archipelago have not yet been fully analyzed, we have to corroborate models of prehistoric human movement using not only new genetic data but also linguistic and archeological data to reconstruct a more comprehensive history of the Japanese people.
Abstract of Paper 6:
Ancient DNA analysis became paleogenomics once high-throughput sequencing technology was applied to ancient DNA sequencing. Paleogenomics based on whole-genome information from Neanderthals and Denisovans showed that small fragments of these genomes remain in the modern human genome, and corresponding studies of anatomical modern humans clarified the history of migration and expansion among Homo sapiens. Due to geographical and environmental conditions, paleogenomic studies have fallen behind in Eastern compared with Western Eurasia. Recently, however, various capture sequencing techniques, which can enrich ancient DNA, have been used in East Eurasia, and the field of paleogenomics has been further developed. This review briefly introduces the history of ancient DNA analysis leading to paleogenomics, outlines three sequencing stages (partial, draft, and complete genome sequencing) and capture methods, and discusses the necessity of high-quality sequencing for paleogenomes of Eastern Eurasia.
The regulator of bone metabolism 〜CC chemokine ligand 28 (CCL28)
Chemokine ligand 28 (CCL28) negatively regulates trabecular bone mass by suppressing osteoblast and osteoclast activities
Rina Iwamoto, Takumi Takahashi, Kazuto Yoshimi, Yuji Imai, Tsuyoshi Koide, Miroku Hara, Tadashi Ninomiya, Hiroaki Nakamura, Kazutoshi Sayama, Akira Yukita
Journal of Bone and Mineral Metabolism 2021 March 15 DOI:10.1007/s00774-021-01210-9
Healthy adult bone mass is tightly controlled by regulator of bone formation and resorption (bone metabolism regulator). The disturbance of this regulation causes pathological bone loss, such as osteoporosis. Recently, the relationship between many chemokines and bone has not yet been investigated, although chemokine has been attracting attention as a bone metabolism regulator.
We performed experiments to clarify the role of CCL28 in bone metabolism, because CCR3 which receptor for CCL28, regulates bone metabolism.
Analysis of Ccl28 deficient mice bone tissue and cultured cell experiments revealed that CCL28 negatively regulates bone mass via suppresses the activation of osteoblasts and osteoclasts. Interestingly, we revealed that Ccl28 expression increased in aged mice bone tissue.
These results provide important insights into bone immunology and the selection of new osteoporosis treatments.
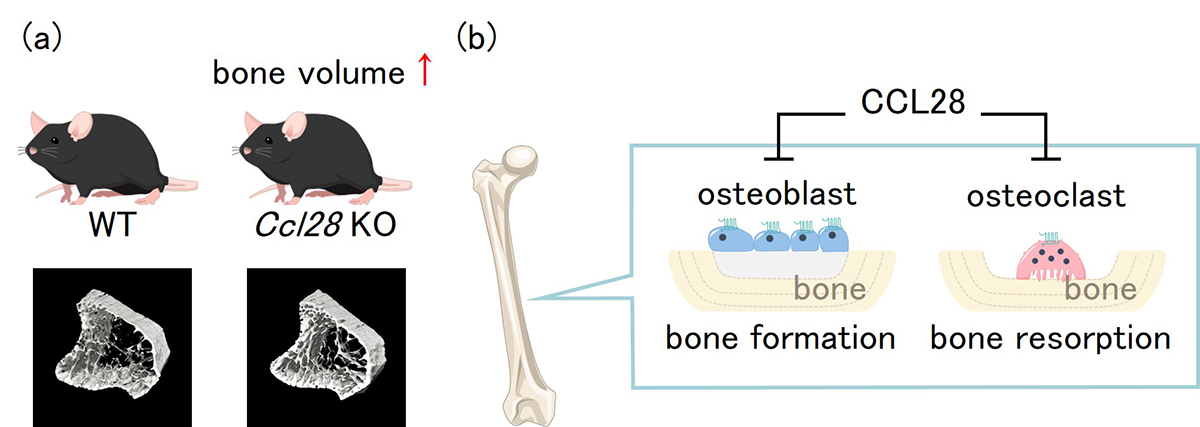
Figure: (a)μCT analysis of bone structure revealed that Ccl28 deficiency increased trabecular bone mass. (b)In vivo and In vitro experiments revealed that CCL28 negatively regulates bone mass by suppressing the activation of osteoblasts and osteoclasts in bone tissue.
Illuminating the role of synapsis in subtelomeric recombination
Sycp1 Is Not Required for Subtelomeric DNA Double-Strand Breaks but Is Required for Homologous Alignment in Zebrafish Spermatocytes
Yukiko Imai, Kenji Saito, Kazumasa Takemoto, Fabien Velilla, Toshihiro Kawasaki, Kei-ichiro Ishiguro and Noriyoshi Sakai
Front Cell Dev Biol 9, 664377 (2021) DOI:10.3389/fcell.2021.664377
Meiosis is a specialized cell division, by which haploid gametes are produced from diploid cells for sexual reproduction. In meiosis, recombination mediates reciprocal exchanges of genetic contents between homologous chromosomes. In human and zebrafish males, meiotic recombination preferentially occurs near telomeres, but the mechanism underlaying the subtelomeric bias remains unknown.
In this study, we investigated roles of physical connection between homologous chromosomes (synapsis) in subtelomeric recombination by using zebrafish. We found that recombination begins near telomeres and homologous chromosomes pair locally, in synapsis-defective zebrafish. However, in the mutant zebrafish, a substantial proportion of pairing was lost in later stages, and no spermatozoa was generated. These observations revealed that synapsis is not essential for initiation of recombination at subtelomeres but is required for its completion.
Since the subtelomeric bias of recombination is also observed in human, new insights into zebrafish meiosis will facilitate understanding human infertility caused by meiotic defects.
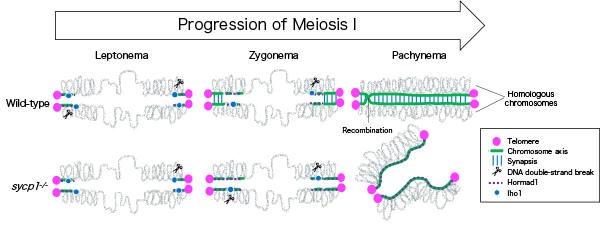
Figure: In wild-type zebrafish males, meiotic recombination begins with DNA double-strand breaks near telomeres. Synapsis is also initiated near telomeres at zygonema, and homologous chromosomes are paired up at pachynema. In sycp1 mutant males, DNA double-strand breaks and local and transient homologous pairing occur near telomeres.
Illuminating ALS Motor Neurons With Optogenetics in Zebrafish
Illuminating ALS Motor Neurons With Optogenetics in Zebrafish
Kazuhide Asakawa, Hiroshi Handa, Koichi Kawakami
Frontiers in Cell Developmental Biology 9, 640414 (2021) DOI:10.3389/fcell.2021.640414
Amyotrophic lateral sclerosis (ALS) is a fatal neurological disorder characterized by progressive degeneration of motor neurons in the brain and spinal cord. Spinal motor neurons align along the spinal cord length within the vertebral column, and extend long axons to connect with skeletal muscles covering the body surface. Due to this anatomy, spinal motor neurons are among the most difficult cells to observe in vivo. Larval zebrafish have transparent bodies that allow non-invasive visualization of whole cells of single spinal motor neurons, from somas to the neuromuscular synapses. This unique feature, combined with its amenability to genome editing, pharmacology, and optogenetics, enables functional analyses of ALS-associated proteins in the spinal motor neurons in vivo with subcellular resolution. Here, we review the zebrafish skeletal neuromuscular system and the optical methods used to study it. We then introduce a recently developed optogenetic zebrafish ALS model that uses light illumination to control oligomerization, phase transition and aggregation of the ALS-associated DNA/RNA-binding protein called TDP-43. Finally, we will discuss how this disease-in-a-fish ALS model can help solve key questions about ALS pathogenesis and lead to new ALS therapeutics.
This review article is a collaborative work conducted by Tokyo Medical University and NIG, and supported by the SERIKA FUND and KAKENHI (JP19K06933 and JP20H05345).
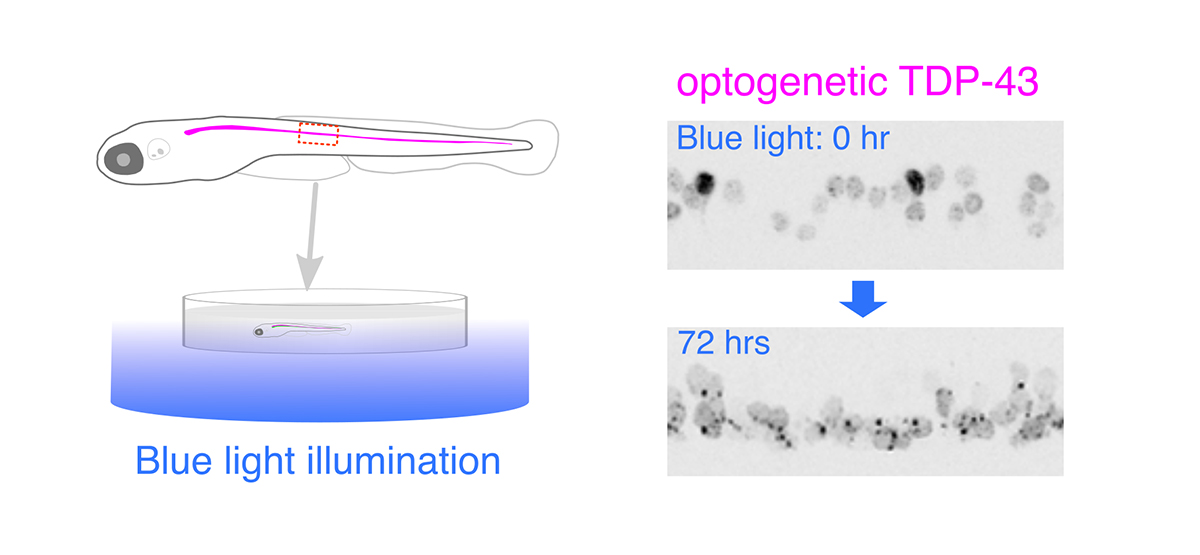
Figure: Optogenetic stimulation of TDP-43 phase transition in the zebrafish spinal motor neurons. Optogenetic TDP-43 forms abnormal aggregates by blue light illumination.
Restoration: Seminar/Meeting system is currently unavailable
The service has been restored as of April 6, 13:00.
Physical nature of chromatin in the nucleus
Physical nature of chromatin in the nucleus
Kazuhiro Maeshima, Shiori Iida, and Sachiko Tamura
Cold Spring Harbor Perspectives in Biology The Nucleus (2nd edition) 13, a040675 (2021) DOI:10.1101/cshperspect.a040675
The full text of the article is here.
Genomic information is encoded on long strands of DNA, which are folded into chromatin and stored in a tiny nucleus. Nuclear chromatin is a negatively charged polymer composed of DNA, histones, and various nonhistone proteins. Because of its highly charged nature, chromatin structure varies greatly depending on the surrounding environment (e.g., cations, molecular crowding, etc.). New technologies to capture chromatin in living cells have been developed over the past ten years. Our view on chromatin organization has drastically shifted from a regular and static one to a more variable and dynamic one. Chromatin forms numerous compact dynamic domains that act as functional units of the genome in higher eukaryotic cells and locally appears liquid-like. Chromatin dynamics can be modulated by several physical or geometrical factors in the cell including cations, chromatin proteins, and transcription machinery. This dynamic behavior can govern various genome functions by changing DNA accessibility. Based on new evidences from versatile genomics and advanced imaging studies, we discuss the physical nature of chromatin in the crowded nuclear environment and how it is regulated. This review article was published in Cold Spring Harbor Perspectives in Biology, also as a chapter of “The Nucleus (2nd edition)”.
This work was supported by JSPS and MEXT KAKENHI grants (20H05936), a Japan Science and Technology Agency CREST grant (JPMJCR15G2), the Takeda Science Foundation, and the Uehara Memorial Foundation.
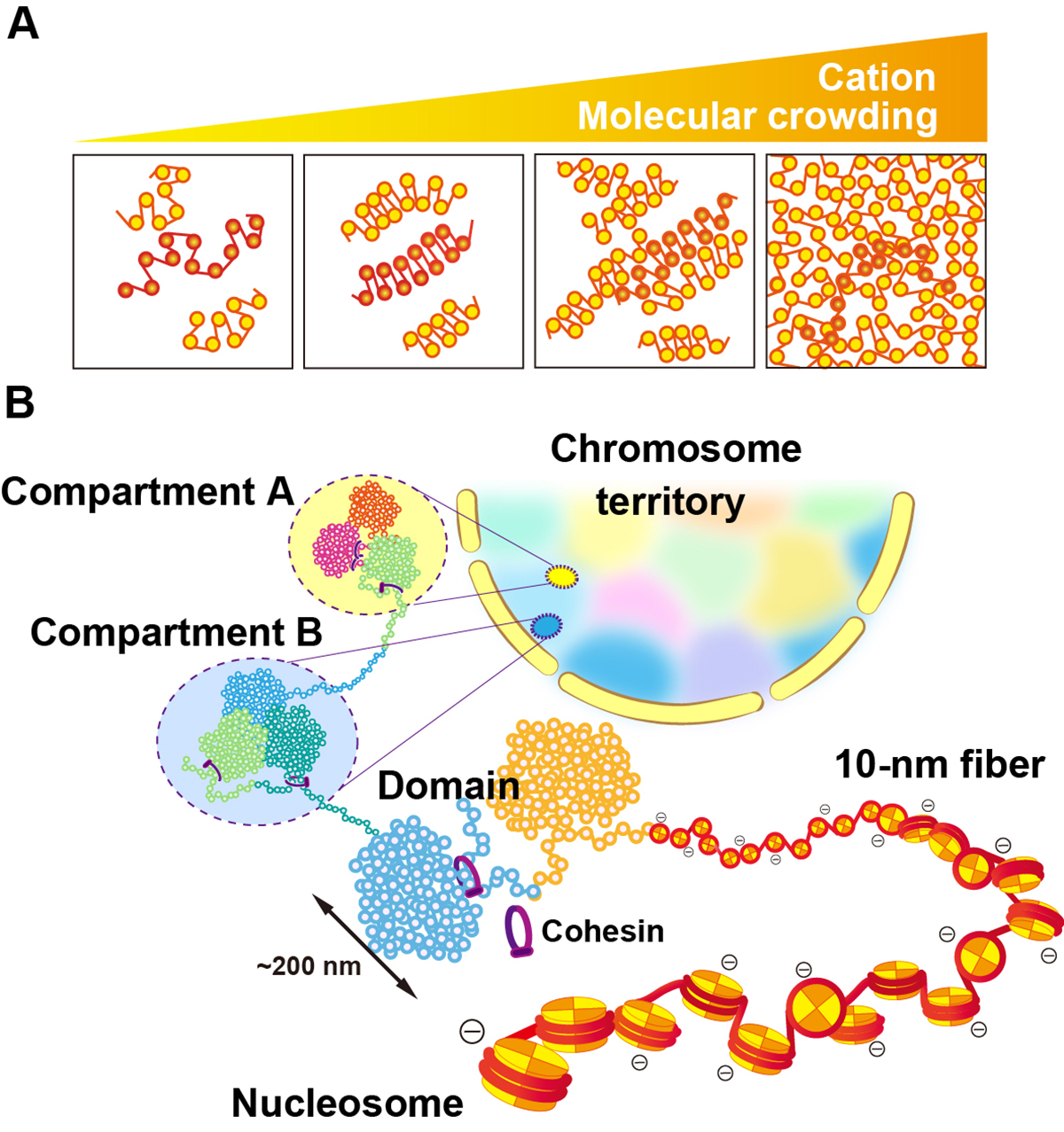
Figure: (A) Polymer melt model. Under physiological cation and/or molecular crowding conditions, inter-fiber nucleosomal contacts in the fibers increase and lead to a melted polymer-like or “sea of nucleosomes” state. (B) A simplified scheme for hierarchical chromatin organization in the nucleus: Nucleosome, the 10-nm fiber, chromatin domains, chromatin compartments and interphase chromosome occupied in a chromosome territory (highlighted as different colors).
Possible roles for DSCR4 (Down Syndrome Critical Region 4) gene
Possible roles for the hominoid-specific DSCR4 gene in human cells
Morteza M. Saber, Marziyeh Karimiavargani, Takanori Uzawa, Nilmini Hettiarachchi, Michiaki Hamada, Yoshihiro Ito and Naruya Saitou
Genes and Genetic Systems 2021 March 24 DOI:10.1266/ggs.20-00012
00012 Down syndrome in humans is caused by trisomy of chromosome 21. DSCR4 (Down syndrome critical region 4) is a de novo-originated protein-coding gene present only in human chromosome 21 and its homologous chromosomes in apes. Despite being located in a medically critical genomic region and an abundance of evidence indicating its functionality, the roles of DSCR4 in human cells are unknown. We used a bioinformatic approach to infer the biological importance and cellular roles of this gene. Our analysis indicates that DSCR4 is likely involved in the regulation of interconnected biological pathways related to cell migration, coagulation and the immune system. We also showed that these predicted biological functions are consistent with tissue-specific expression of DSCR4 in migratory immune system leukocyte cells and neural crest cells (NCCs) that shape facial morphology in the human embryo. The immune system and NCCs are known to be affected in Down syndrome individuals, who suffer from DSCR4 misregulation, which further supports our findings. Providing evidence for the critical roles of DSCR4 in human cells, our findings establish the basis for further experimental investigations that will be necessary to confirm the roles of DSCR4 in the etiology of Down syndrome.
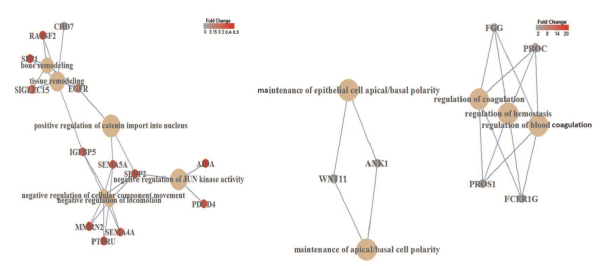
Figure: Gene regulatory network analysis of DSCR4 overexpression-mediated diffentially expressed genes. Left: diffentially expressed genes down-regulated by DSCR4 overexpression revealed enrichment of six interconnected pathways involved in the regulation of migration, locomotion and remodeling processes. Right: up-regulated diffentially expressed genes showed enrichment for three interconnected pathways involved in the regulation of blood coagulation and hemostasis along with maintenance of apical/basal cell polarity.
New faculty appointments as of April 1st
Dr. Shigehiro Kuraku joined NIG as a professor on April 1.
KURAKU, Shigehiro : Molecular Life History Laboratory

- KURAKU, Shigehiro professor
Dr. Hiroshi Mori opened a new laboratory as an associate professor on April 1.
MORI, Hiroshi Associate Professor : Advanced Genomics Center / Genome Diversity Laboratory
- MORI, Hiroshi Associate Professor
Two assistant professors joined NIG.
SAITO, Kei : Shimamoto Group • Physics and Cell Biology Laboratory















