Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
A novel antitumor platinum complex
Biological Macromolecules Laboratory / Maeshima Group
Chromatin folding and DNA replication inhibition mediated by a highly antitumor-active tetrazolato-bridged dinuclear platinum(II) complex
Ryosuke Imai, Seiji Komeda, Mari Shimura, Sachiko Tamura, Satoshi Matsuyama, Kohei Nishimura, Ryan Rogge, Akihiro Matsunaga, Ichiro Hiratani, Hideaki Takata, Masako Uemura, Yutaka Iida, Yuko Yoshikawa, Jeffrey C. Hansen, Kazuto Yamauchi, Masato T. Kanemaki, and Kazuhiro Maeshima
Scientific Reports 6, Article number: 24712 (2016) DOI:10.1038/srep24712
Chromatin DNA must be read out for various cellular functions, and copied for the next cell division. These processes are targets of many anticancer agents. Platinum-based drugs, such as cisplatin, have been used extensively in cancer chemotherapy. The drug–DNA interaction causes DNA crosslinks and subsequent cytotoxicity. Recently, it was reported that an azolato-bridged dinuclear platinum (II) complex, 5-H-Y, exhibits a different anticancer spectrum from cisplatin. Here, using an interdisciplinary approach, we reveal that the cytotoxic mechanism of 5-H-Y is distinct from that of cisplatin. 5-H-Y inhibits DNA replication and also RNA transcription, arresting cells in the S/G2 phase, and are effective to cisplatin-resistant cancer cells. Moreover, it causes much less DNA crosslinking than cisplatin, and induces chromatin folding. 5-H-Y will expand the clinical applications for the treatment of chemotherapy-insensitive cancers.
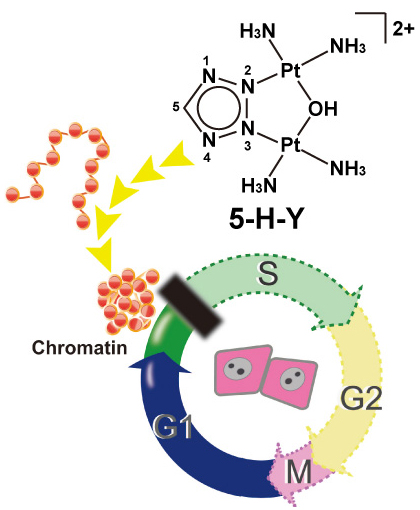
5-H-Y inhibits DNA replication and arrests the treated cells in S/G2 phase. 5-H-Y binds tightly to chromatin DNA and induces chromatin folding in vitro and in vivo.
Genomic locations of conserved noncoding sequences and relationship with their function
Division of Population Genetics / Saitou Group
Genomic locations of conserved noncoding sequences and their proximal protein-coding genes in mammalian expression dynamics
Isaac Adeyemi Babarinde and Naruya Saitou
Molecular Biology and Evolution DOI:10.1093/molbev/msw058
Conserved Noncoding Sequences (CNSs) in mammals were examined with special reference to their genomic location. We extracted the CNSs conserved between chicken and four mammalian species (human, mouse, dog and cattle). Human CNSs were confirmed to be under purifying selection. The distribution pattern, ChIP-Seq and RNA-Seq data suggested that the CNSs are regulatory elements. Physical distances between CNS and their nearest protein coding genes were well conserved between human and mouse genomes. ChIP-Seq signal and gene expression patterns also suggested that CNSs regulate nearby genes. Genes with more CNSs have more evolutionarily conserved expression than those with fewer CNSs. These results suggest that the genomic locations of CNSs are important for their regulatory functions. In fact, various kinds of evolutionary constraints may be acting to maintain the genomic locations of CNSs and protein-coding genes in mammals to ensure proper regulation. First author of this paper, Dr. Babarinde, just receive Ph.D. from Department of Genetics, SOKENDAI. He also received Morishima Award from NIG.
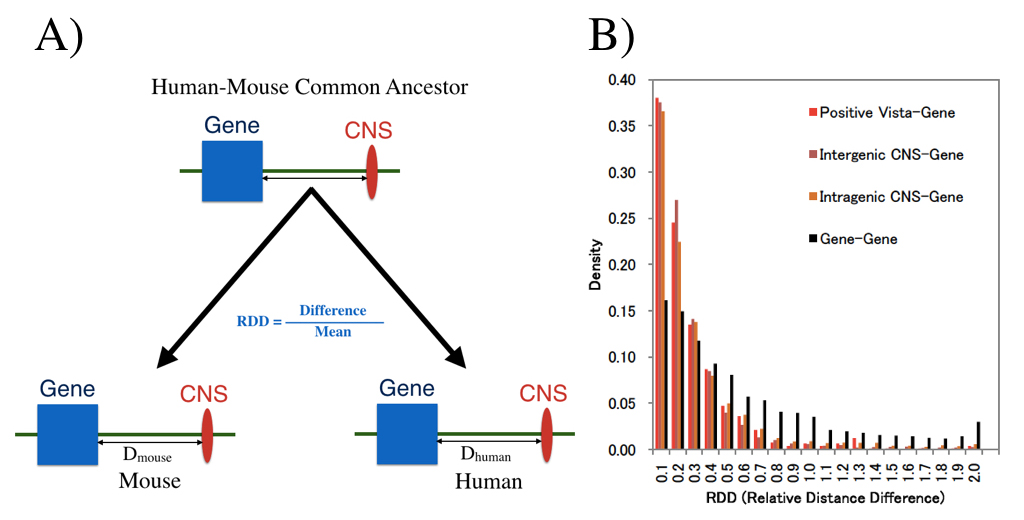
We examined whether genomic distances between genes and CNSs which already existed in the common ancestor of human and house are conserved now. (A) shows definition of distance measure RDD, and (B) shows RDD distribution. Distances between CNS and genes are much more conserved than those between genes.
Dazl is a target RNA suppressed by mammalian NANOS2 in sexually differentiating male germ cells
Press release
Dazl is a target RNA suppressed by mammalian NANOS2 in sexually differentiating male germ cells
Yuzuru Kato, Takeo Katsuki, Hiroki Kokubo, Aki Masuda, Yumiko Saga
Nature Communications 7, Article number: 11272 DOI:10.1038/NCOMMS11272
Press Release (Only in Japanese)
Evolutionally conserved Nanos RNA-binding proteins play crucial roles in germ cell development. While a mammalian Nanos family protein, NANOS2, is required for sexual differentiation of male (XY) germ cells in mice, the underlying mechanisms and the identities of its target RNAs in vivo remain elusive. Using comprehensive microarray analysis and a bacterial artificial chromosome transgenic system, we identify Dazl, a germ cell-specific gene encoding an RNA-binding protein implicated in translation, as a crucial target of NANOS2. Importantly, removal of the Dazl 3′ UTR in XY germ cells stabilises the Dazl mRNA, resulting in elevated meiotic gene expression, abnormal resumption of the cell cycle, and impaired processing-body formation, reminiscent of Nanos2-knockout phenotypes. Furthermore, our data suggest that NANOS2 acts as an antagonist of the DAZL protein. We propose a dual system of NANOS2-mediated suppression of Dazl expression as a pivotal molecular mechanism promoting sexual differentiation of XY germ cells. This work was supported by a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (JSPS) to Y.K. (Grant number 25840091) and a Grant-in-Aid for Scientific Research (S) from JSPS to Y.S. (Grant number 21227008).
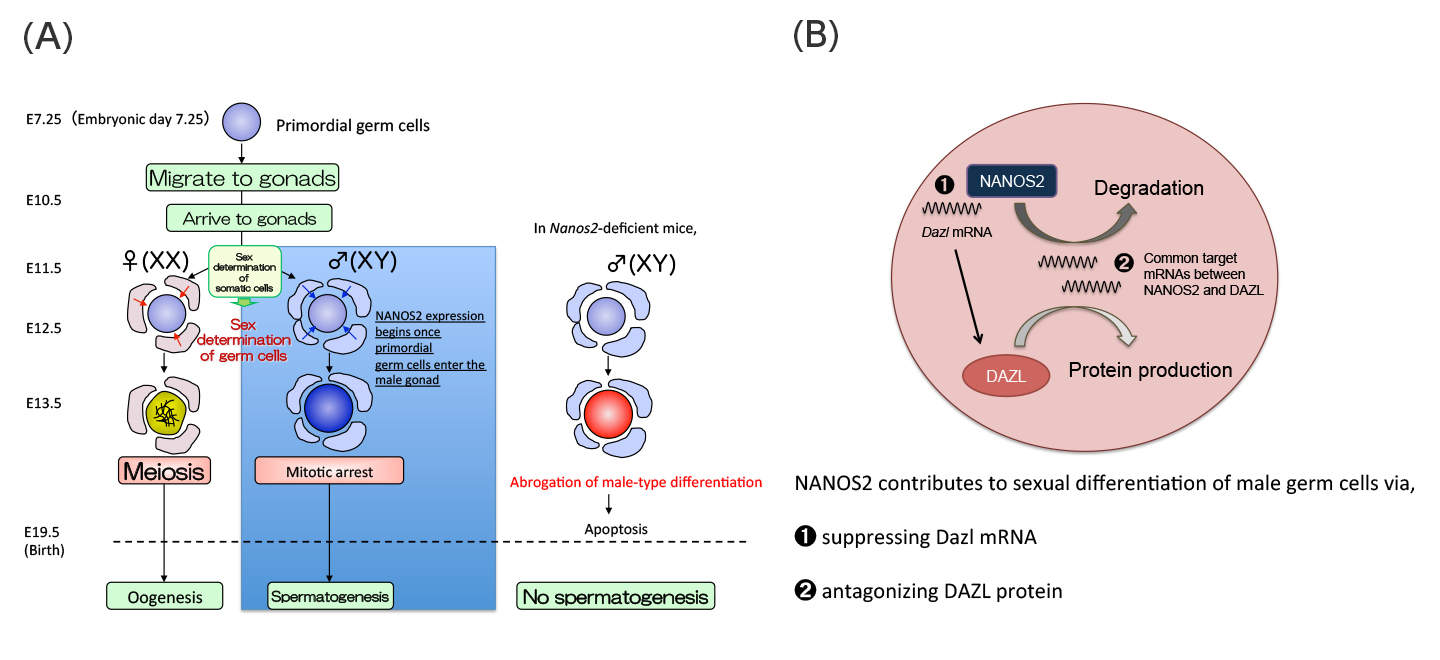
(A)Sex of mouse germ cells is determined after they are enclosed by gonadal somatic cells at E11.5. NANOS2 expression begins once primordial germ cells enter the sex-specific process of sexual differentiation in male germ cells. Germ cells lacking the Nanos2 gene die by apoptosis due to the abrogated male-type differentiation.
(B)Working model of NANOS2-dependent Dazl suppression in sexually differentiating male germ cells.
Genome chromatin has an intrinsic property of irregular folding.
Press release
Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers
Kazuhiro Maeshima, Ryan Rogge, Sachiko Tamura, Yasumasa Joti, Takaaki Hikima, Heather Szerlong, Christine Krause, Jake Herman, Erik Seidel, Jennifer DeLuca, Tetsuya Ishikawa, and Jeffrey C. Hansen
The EMBO Journal Published online 12.04.2016 DOI:10.15252/embj.201592660
The existence of a 30-nm fiber as a basic folding unit for DNA packaging has remained a topic of active discussion. Here we characterize the supramolecular structures formed by reversible Mg2+-dependent self-association of linear 12-mer nucleosomal arrays using microscopy and physicochemical approaches. These structures, which we call ’oligomers’, were globular throughout all stages of the cooperative assembly process, and ranged in size from ~50 nm to a maximum diameter of ~1000 nm. The nucleosomal arrays were packaged within the oligomers as interdigitated 10-nm fibers, rather than folded 30-nm structures. Linker DNA was freely accessible to micrococcal nuclease, although the oligomers remained partially intact after linker DNA digestion. The organization of chromosomal fibers in human nuclei in situ was stabilized by 1 mM MgCl2 but became disrupted in the absence of MgCl2, conditions that also dissociated the oligomers in vitro. These results indicate that a 10-nm array of nucleosomes has the intrinsic ability to self-assemble into large chromatin globules stabilized by nucleosome-nucleosome interactions, and suggest that the oligomers are good in vitro model for investigating the structure and organization of interphase chromosomes.
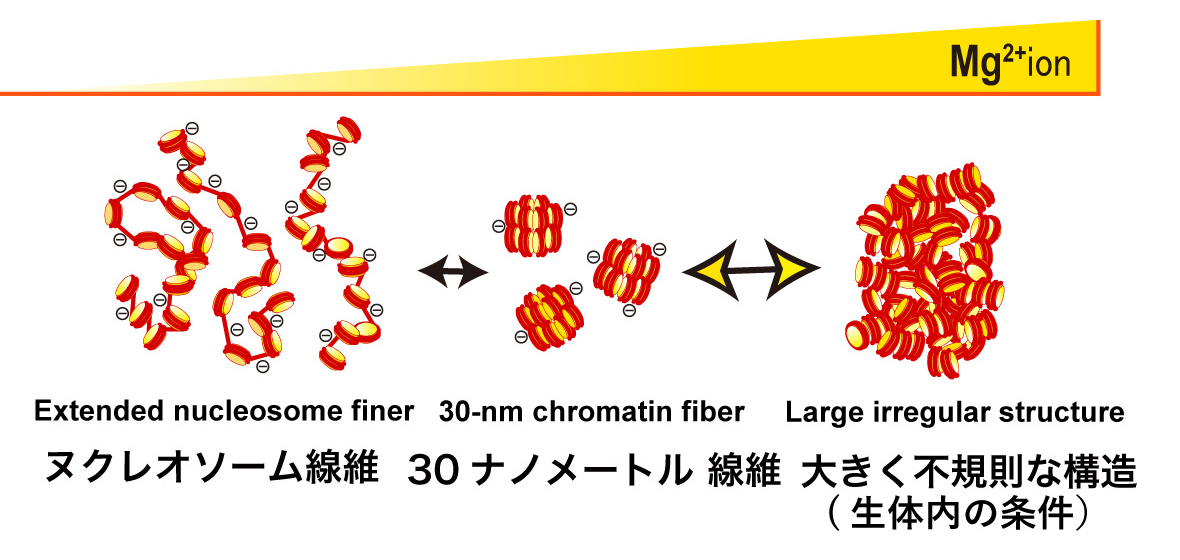
The 12-mer nucleosomal array is a well-defined model chromatin system. Without Mg2+, the nucleosomal array is extended by repulsion (Left). In 1-2 mM Mg2+, the nucleosomal array folds into a regular 30-nm chromatin fiber structure (Center). With further increases in Mg2+, the nucleosome arrays assemble into supramolecular structures (Right). The large structures are not assemblies of the 30-nm chromatin fibers, but interdigitated and melted structures of 10-nm nucleosomal arrays, reflecting on the native chromatin structure in the cell.
Allelic imbalance in regulation of ANRIL through chromatin interaction at 9p21 endometriosis risk locus
Press release
Allelic Imbalance in Regulation of ANRIL through Chromatin Interaction at 9p21 Endometriosis Risk Locus
Hirofumi Nakaoka, Aishwarya Gurumurthy, Takahide Hayano, Somayeh Ahmadloo, Waleed H Omer, Kosuke Yoshihara, Akihito Yamamoto, Keisuke Kurose, Takayuki Enomoto, Shigeo Akira, Kazuyoshi Hosomichi, Ituro Inoue
PLOS Genetics Published: April 7, 2016 DOI:10.1371/journal.pgen.1005893
Press Release (Only in Japanese)
A large number of variants associated with human complex diseases have been discovered by genome-wide association studies (GWASs). These discoveries have been anticipated to be translated into the definitive understanding of disease pathogeneses; however, functional characterization of the disease-associated SNPs remains a formidable challenge. Here we explored regulatory mechanism of a variant on chromosome 9p21 associated with endometriosis, a common gynecological disorder. By scrutinizing linkage disequilibrium structure and DNase I hypersensitive sites across the risk locus, we prioritized rs17761446 as a candidate causal variant. The results of our “allele-specific” functional genomic approaches sheds light on regulatory mechanisms underlying 9p21 endometriosis risk locus, in which preferential bindings of TCF7L2 and its coactivator EP300 to the protective G allele of rs17761446 lead to stronger chromatin interaction with the promoter of ANRIL, which in turn activate transcription of the non-coding RNA. Motivated by the fact that TCF7L2 was a key transcription factor of Wnt signaling pathway, we postulated that the induction of Wnt signaling activated expression levels of ANRIL and cell cycle inhibitors, CDKN2A/2B. Functional genomics on common disease will unlock functional aspect of genotype-phenotype correlations in the post-GWAS stage.
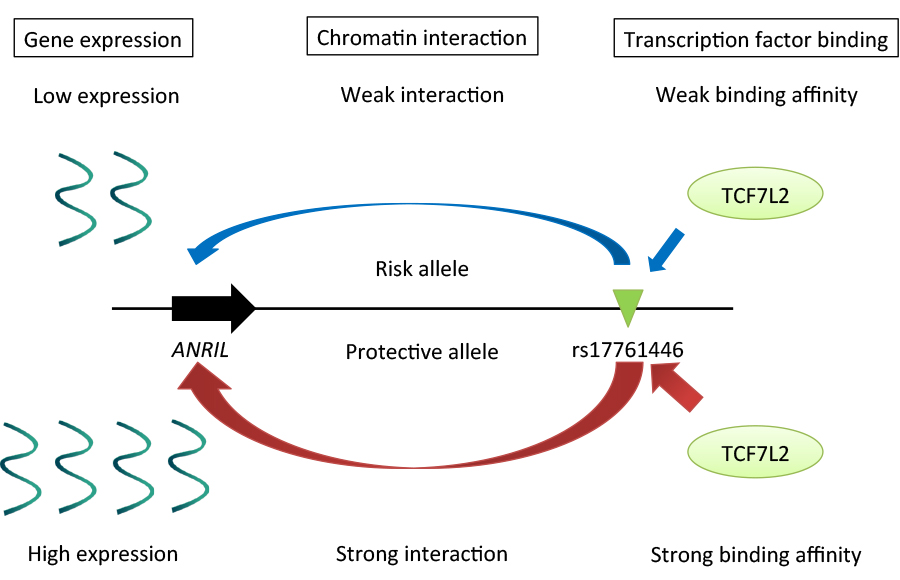
Regulatory mechanisms underlying 9p21 endometriosis risk locus. Preferential bindings of TCF7L2 and its coactivator EP300 to the protective G allele of rs17761446 lead to stronger chromatin interaction with the promoter of ANRIL, which in turn activate transcription of the non-coding RNA.
The mechanism of centering of the cell nucleus inside the cell
Cell Architecture Laboratory / Kimura Group
Shape–motion relationships of centering microtubule asters
Hirokazu Tanimoto, Akatsuki Kimura*, and Nicolas Minc*
J. Cell Biol. 212: 777-787, 2016. DOI:10.1083/jcb.201510064
(* corresponding authors)
A joint group of Drs. Nicolas Minc at Institut Jacques Monod (France) and Akatsuki Kimura at National Institute of Genetics/SOKENDAI analyzed the centering of microtubule asters in the sea urchin egg, and constructed a quantitative model that dictates the speed and trajectory of the centering in normal cell as well as shape-manipulated cells. The study was initiated when Prof. Kimura was a visiting researcher at the Minc lab through the SOKENDAI Young Faculty Overseas Visit Program.
In this study, Dr. Hirokazu Tanimoto et al. utilized multiple approaches such as laser ablation and pharmacological assays to demonstrate that the centering is accomplished by the forces pulling on asters from the cytoplasm. An interesting feature of the centering was to find that asters moved at a constant speed. Considering the speed was comparable to that of the growth speed of the aster, Tanimoto et al. proposed a theoretical model for the centering that explains the constant speed independent on the actual size of the aster. The model predicts well the change of the speed and trajectory of asters when the shape of the cells was manipulated in micro-chambers. Overall, this study advanced our understanding on the long-standing debate on the mechanism of the centering of microtubule asters and their associating nucleus.
See also: Ben Short “How asters find their center” J Cell Biol 212, 743 (2016) DOI: 10.1083/jcb.2127if
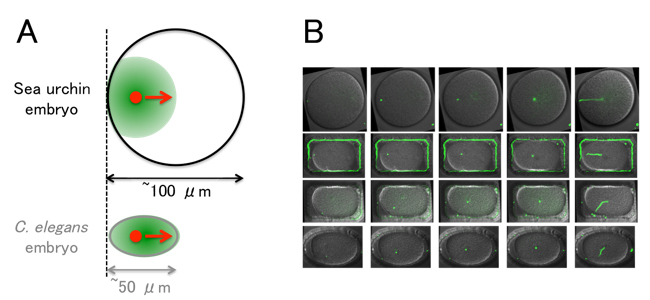
(A) The centering of microtubule asters (green) in the sea urchin embryo. The size of the embryo is large compared to C. elegans. The aster does not cover the entire cell during the centering, and the center of the aster (red) approaches the cell center with a speed comparable to the growth speed of the aster.
(B) Examples of shape manipulation using micro-chambers. The sperm pronucleus (i.e. the center of the aster, green) moves toward the center. (The entire trajectories are shown on the rightmost panels.) The theoretical model constructed in the study reproduces the speed and trajectory of aster movements even in the shape-manipulated embryos.
OPEN HOUSE 2016
Time & Date
9:00 ~ 16:00, Saturday, April 2th, 2016 (No Reservations Required, Free Admission)
Access
Free Shuttle Buses Available to the National Institute of Genetics from the North Exit of Mishima Station. ( Service Time: 8:50 ~ 15:00)
Map
Map Free Shuttle Buses Available Parking around Mishima Station Available for Car Visitors.
No Pets Allowed except Service Dogs, No General Parking*
(*Disabled Parking Available on the Institute Premises)
Information
1111 Yata, Mishima, Shizuoka 411-8540, JAPAN TEL:+81-55-981-5873
New faculty appointments and tenure promotion as of April 1st
Two professors and three assistant professors have joined NIG as of April 1, 2016.
professor
Yutaka SATO:Genetic Resource Center,Plant Genetics Laboratory
Ken KUROKAWA:Center for Information Biology, Genome Evolution Laboratory
- Yutaka SATO
professor

- Ken KUROKAWA
professor
assistant professor
Takayuki FUJIWARA: Division of Symbiosis and Cell Evolution, Miyagishima Group
Tomotaka MATSUMOTO: Division of Evolutionary Genetics, Akashi Group
Toshihiro KAWASAKI: Model Fish Genomics Resource Laboratory, Sakai Group
In addition, Masato KANEMAKI, tenure track associate professor at the Center for Frontier Research, has been awarded tenure and promoted to professor.
Masato KANEMAKI: Division of Molecular Cell Engineering, Department of Molecular Genetics

- Masato KANEMAKI
professor















