Revealing the shared features of “Hard-to-synthesize” proteins in Bacteria — and discovering unique protein groups that actively exploits them
Press release
Evolutionary Adaptation of Bacterial proteomes to Translation-Impeding Sequences
Keigo Fujiwara, Naoko Tsuji, Karen Sakiyama, Hironori Niki, and Shinobu Chiba
The EMBO Journal (2025)
![]() Press release (In Japanese only)
Press release (In Japanese only)
In this study, we revealed that diverse bacteria on Earth share amino acid sequences that are difficult for them to synthesize. Because amino acids are essential building blocks of proteins, cells cannot efficiently produce proteins that contain such translation-challenging sequences. Indeed, a comprehensive analysis conducted across the bacterial domain showed that these translation-challenging sequences are rarely found within bacterial proteins, suggesting that bacteria have evolutionarily avoided using them.
In contrast, we also discovered that these sequences frequently appear near the ends (carboxyl termini) of relatively small proteins. Bioinformatic analyses further indicated that such proteins likely play important and diverse roles in helping cells adapt to fluctuating environmental conditions.
Together, this work demonstrates that translation-challenging sequences, although generally disadvantageous and evolutionarily excluded from most proteins, can be repurposed by bacteria as functional elements. By leveraging the “difficulty” of translating these sequences, various bacteria appear to have evolved unique strategies to better cope with environmental changes.
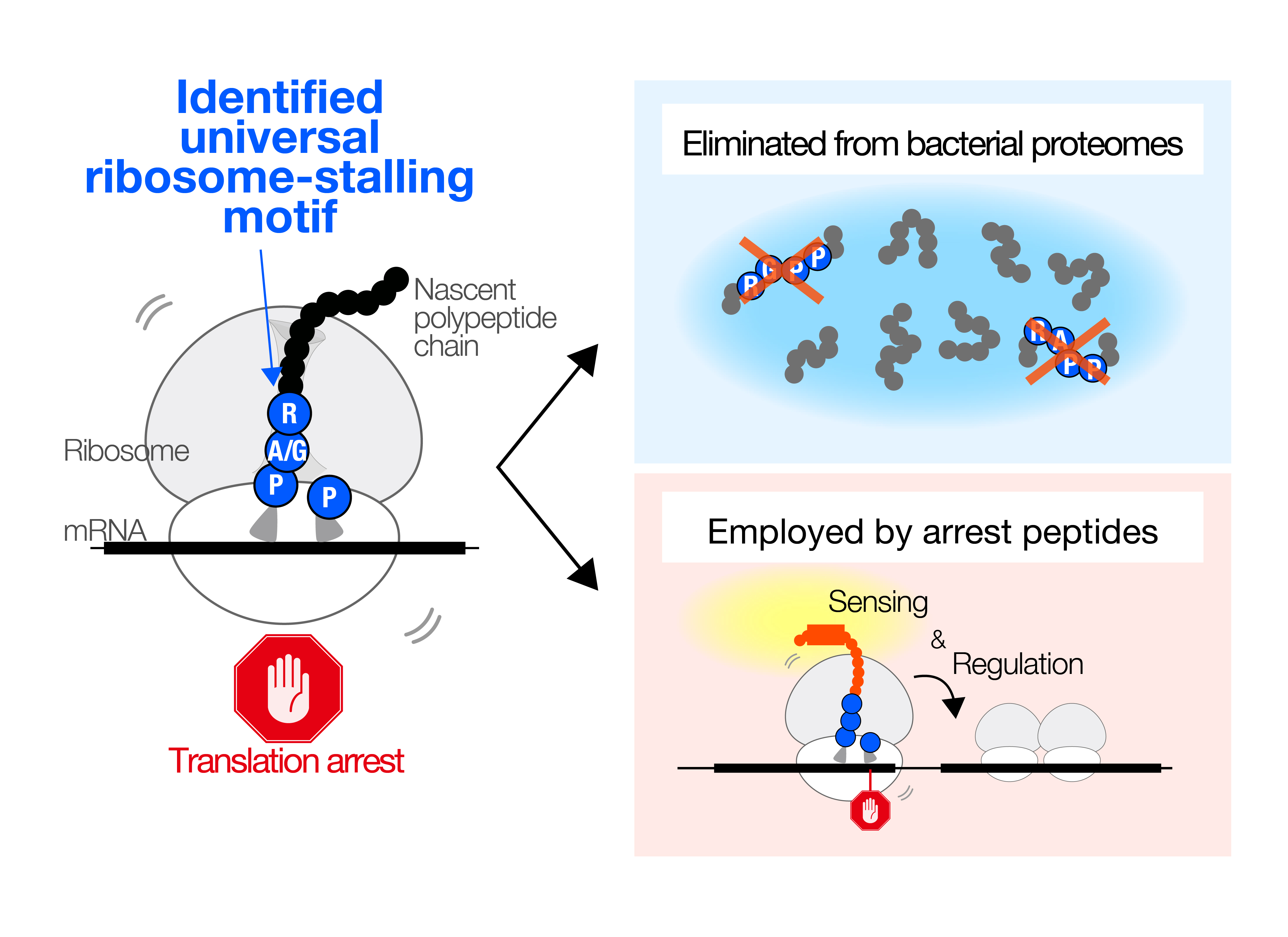
Figure:We identified patterns of “translation arrest-causing amino acid sequences” that occur across many bacteria (left). Although such sequence patterns are generally eliminated during evolution (upper right), some bacteria have instead evolved unique mechanisms that leverage this inherent synthesis difficulty to support important cellular functions (lower right)















