Uncovering the principle by which DNA replication initiation sites are determined in the human genome
Press release
Regulated TRESLIN-MTBP loading governs initiation zones and replication timing in human DNA replication
Xiaoxuan Zhu, Atabek Bektash, Yuki Hatoyama, Sachiko Muramatsu, Shin-Ya Isobe, Chikashi Obuse, Atsushi Toyoda, Yasukazu Daigaku, Chun-Long Chen and Masato T. Kanemaki
Nature Communications 2025 DOI:10.1038/s41467-025-66278-7
![]() Press release (In Japanese only)
Press release (In Japanese only)
When cells proliferate, genomic DNA is precisely duplicated once per cell cycle. Abnormalities in this DNA replication process can cause alterations in genomic DNA, contributing to cellular ageing, cancer, and hereditary diseases. Therefore, understanding how cells replicate their DNA is crucial for elucidating fundamental biological processes, diseases, and even evolution.
Traditionally, DNA replication has been studied in microorganisms such as E. coli and yeast. In these organisms, the locations where DNA replication begins (replication origins) are determined by specific DNA sequences. However, in most eukaryotic cells, including human cells, the DNA sequence itself does not dictate where replication starts. For decades, it remained a mystery how and where replication is initiated within the human genome.
To address this, Professor Masato Kanemaki and his team including the first author, Dr Xiaoxuan Zhu, at National Institute of Genetics developed a new high-precision method, LD-OK-seq (Ligase Depletion-Okazaki sequencing), to detect replication initiation regions in the human genome. By further analysing the proteins bound to these regions, they uncovered the fundamental principle by which human cells determine replication initiation sites.
Their findings revealed that, except for actively transcribed gene regions, human cells possess the ability to initiate DNA replication from almost anywhere in the genome. This capability arises from the widespread binding of an enzyme called the MCM helicase, which is essential for DNA replication. Moreover, they discovered that during the early S phase, replication frequently begins in intergenic regions (areas between transcribed genes), and that these sites are determined by the binding of TRESLIN-MTBP, a protein complex that activates the MCM helicase. They also identified an antagonistic regulatory system that modulates the binding of TRESLIN-MTBP to MCM.
These discoveries answer the fundamental question of how human cells initiate genome replication, providing new insights into diseases caused by replication abnormalities—such as genomic instability disorders, cancer, aging, and hereditary diseases—as well as into genome evolution. In the long term, this work may also lay the foundation for technologies that enable artificial control of DNA replication.
This study was conducted through an international collaboration between the research group of Professor Masato Kanemaki and Project Professor Atsushi Toyoda at the National Institute of Genetics, Professor Chikashi Obuse at The University of Osaka, Dr. Yasukazu Daigaku at the Cancer Institute of JFCR, and Professor Chun-Long Chen at Curie Institute in France. The research was supported by JSPS KAKENHI grants (JP23H02463, JP21H04719, JP23H04925, JP25H00979), Platform for Advanced Genome Science (JP22H04925), JST FOREST (JPMJFR204X), JST CREST (JPMJCR21E6), and AMED ASPIRE (JP25jf0126015).
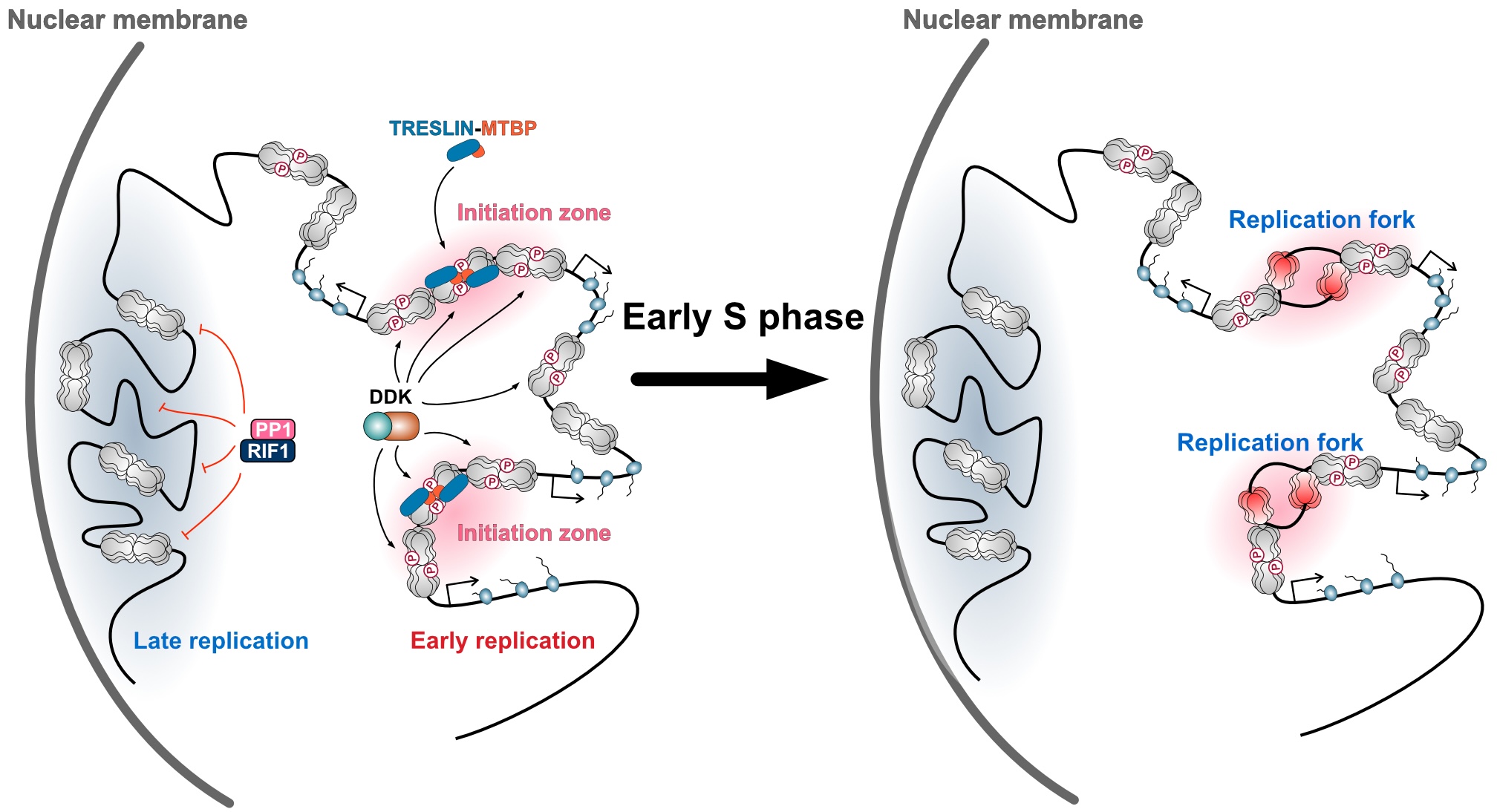
Figure: The MCM helicase is broadly bound across genomic DNA in regions outside actively transcribed genes, and its phosphorylation is antagonistically regulated by the kinase DDK and the phosphatase RIF1–PP1. The sites of replication initiation are determined when the TRESLIN–MTBP complex is recruited to the phosphorylated MCM helicase.















