Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
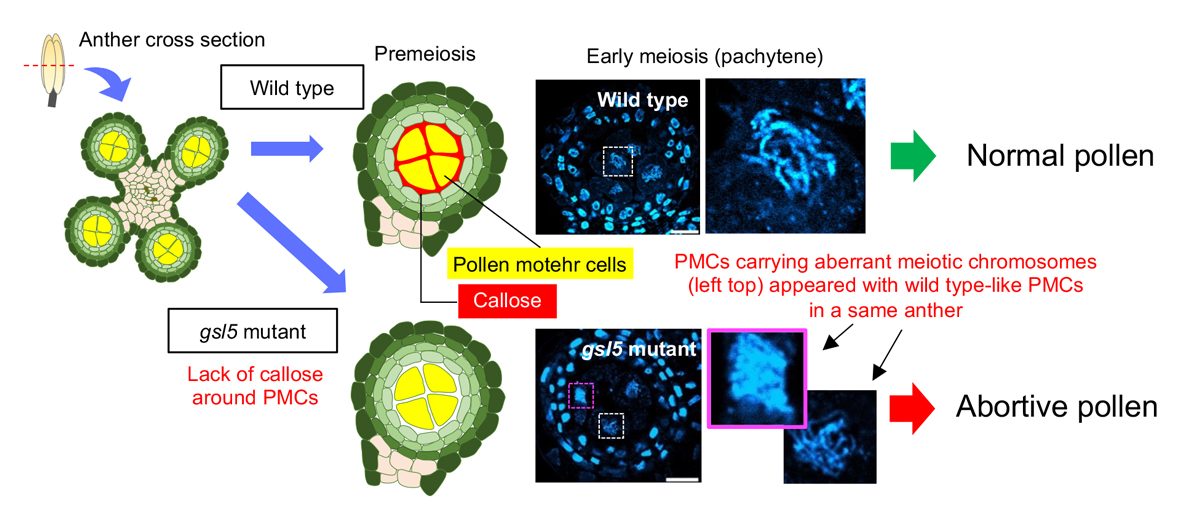
Rice GLUCAN SYNTHASE-LIKE5 promotes anther callose deposition to maintain meiosis initiation and progression
Press release
Rice GLUCAN SYNTHASE-LIKE5 promotes anther callose deposition to maintain meiosis initiation and progression
H. Somashekar, M. Mimura, K. Tsuda, K. I. Nonomura
Plant Physiology 2022 October 22 DOI:10.1093/plphys/kiac488
![]() Press release (In Japanese only)
Press release (In Japanese only)
Callose is a plant cell-wall polysaccharide whose deposition is spatiotemporally regulated in various developmental processes and environmental stress responses. Appearance of callose in premeiotic anthers is a prominent histological hallmark for the onset of meiosis in flowering plants; however, the biological role of callose in meiosis remains unknown. Here we show that rice (Oryza sativa) GLUCAN SYNTHASE LIKE5 (OsGSL5), a callose synthase, localizes on the plasma membrane of pollen mother cells (PMCs) and is responsible for biogenesis of callose in anther locules through premeiotic and meiotic stages. In Osgsl5 mutant anthers mostly lacking callose deposition, aberrant PMCs accompanied by aggregated, unpaired or multivalent chromosomes were frequently observed, and furthermore, a considerable number of mutant PMCs had untimely progress into meiosis compared to that of wild-type PMCs. Immunostaining of meiosis-specific protein HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS2 (PAIR2) in premeiotic PMCs revealed precocious meiosis entry in Osgsl5 mutant anthers. These findings provide insights into the function of callose in controlling the timing of male meiosis initiation and progression, in addition to roles in microsporogenesis, in flowering plants.
Source: H. Somashekar et al., DOI: 10.1093/plphys/kiac488
How to build the cellular machine for error-free chromosome segregation
Press release
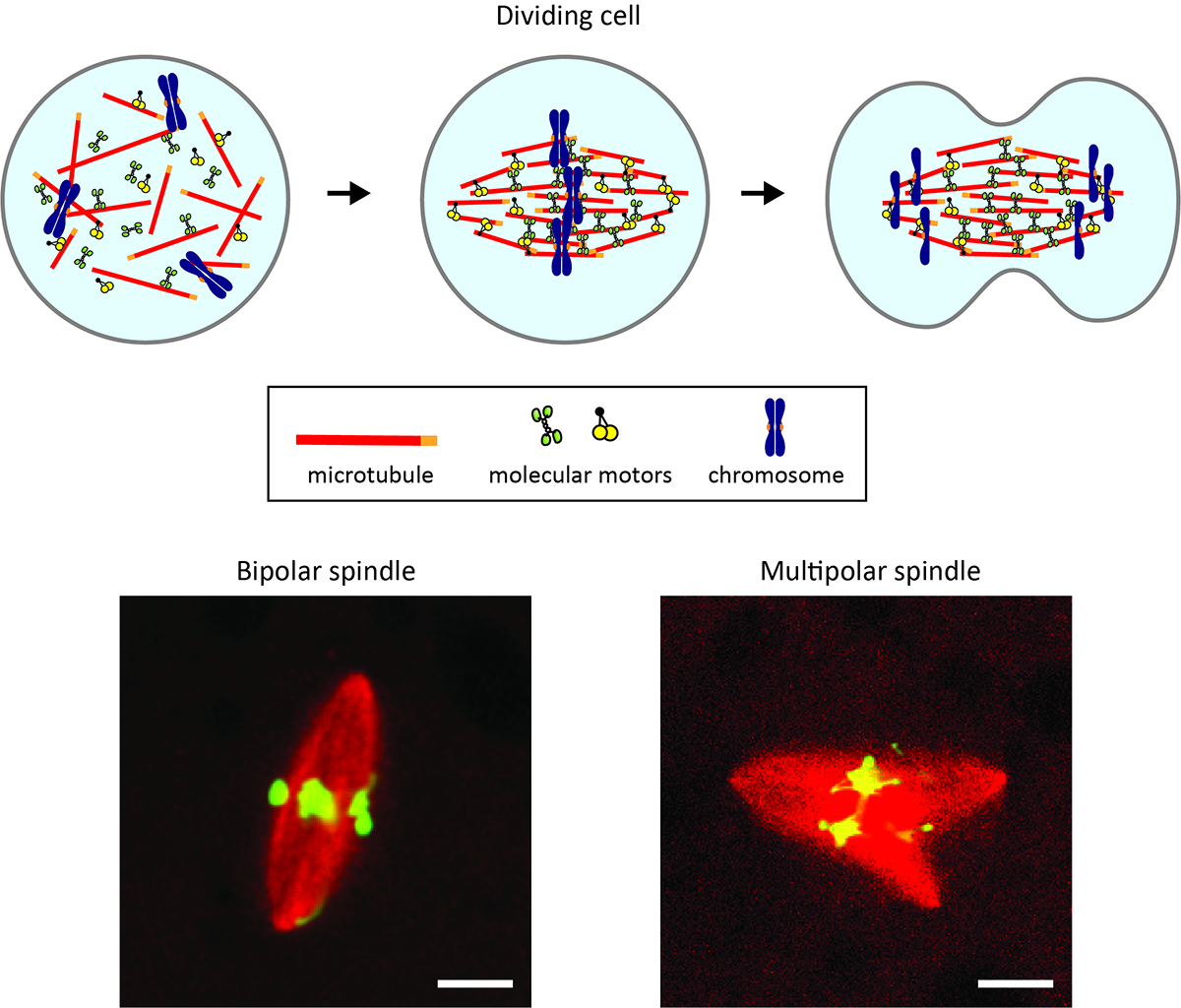
Morphological growth dynamics, mechanical stability, and active microtubule mechanics underlying spindle self-organization
T. Fukuyama, L. Yan, M. Tanaka, M. Yamaoka, K. Saito, SC. Ti, CC. Liao, KC. Hsia, YT. Maeda, Y. Shimamoto
PNAS (2022) 119, e2209053119 DOI:10.1073/pnas.2209053119
![]() Press release (In Japanese only)
Press release (In Japanese only)
Each time a cell divides in our body, it builds a tiny micron-sized machine called the spindle. The spindle passes the copied DNA from a parental cell to the newly-created two daughter cells and should have a bipolar, football-like shape for evenly splitting the chromosomes that carry the DNA. Disrupted spindle shapes can cause errors in chromosome splitting and malfunctions in the cell offspring.
Now, a new study reveals how the shaping of this tiny segregation machine succeeds and when it ends up with failure. A team of biophysics groups in Japan (Yuta Shimamoto’s lab at the National Institute of Genetics and Yusuke Maeda’s lab at Kyushu University) and biochemistry groups abroad (Kuo-Chiang Hsia’s group at Academia Sinica and Jeff Ti’s group at the University of Hong Kong) develop a computer-based algorithm and fluorescence microscopy to track at an unprecedented resolution the way in which the machine is built from scratch. They find the unique “assembly instruction” for making the spindle with the correct shape and one for making the spindle with unfavored shapes. The team also uncovers a memory form-like material property of the spindle, which appears to be a key that separates success from failure in assembling the machine.
Abnormally shaped spindles are linked to conditions that impact human life, such as cancer and infertility. The findings of this study should be an important step toward understanding the relevant mechanisms.
Life cycle and functional genomics of the unicellular red alga Galdieria for elucidating algal and plant evolution and industrial use
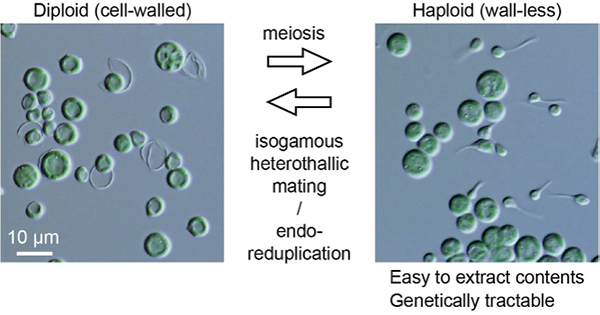
Life cycle and functional genomics of the unicellular red alga Galdieria for elucidating algal and plant evolution and industrial use
S. Hirooka, T. Itabashi, T. M. Ichinose, R. Onuma, T. Fujiwara, S. Yamashita, L. W. Jong, R. Tomita, A. H. Iwane. S. Miyagishima
PNAS (2022) 119, e2210665119 DOI:10.1073/pnas.2210665119
![]() Press release (In Japanese only)
Press release (In Japanese only)
Sexual reproduction is widespread in eukaryotes. However, only asexual reproduction has been observed in unicellular red algae, including Galdieria, which branched early in Archaeplastida. Galdieria possesses a small genome; it is polyextremophile, grows either photoautotrophically, mixotrophically, or heterotrophically and is being developed as an industrial source of vitamins and pigments because of its high biomass productivity. Here, we show that Galdieria exhibits a sexual life cycle, alternating between cell-walled diploid and cell wall-less haploid and that both phases can proliferate asexually. The haploid can move over surfaces and undergo self-diploidization or generate heterozygous diploids through mating. Further, we have prepared the whole genome and comparative transcriptome dataset between the diploid and haploid and developed genetic tools for the stable gene expression, gene disruption and selectable marker recycling system using the cell wall-less haploid. The BELL/KNOX and MADS-box transcription factors, which function in haploid-diploid transition and development in plants, are specifically expressed in the haploid and diploid, respectively, and are involved in the haploid-diploid transition in Galdieria, providing information on the missing link of the sexual life cycle evolution in Archaeplastida. Four actin genes are differently involved in motility of the haploid and cytokinesis in the diploid both of which are myosin-independent and likely reflect ancestral roles of actin. We have also generated photosynthesis-deficient mutants such as blue-colored cells, which were depleted in chlorophyll and carotenoids, for industrial pigment production. These features of Galdieria facilitate understanding the evolution of algae and plants and the industrial use of microalgae.
Source: S. Hirooka et al., PNAS DOI: 10.1073/pnas.2210665119
A review by Professor Ursula Goodenough (Washington University in St. Louis) about this article will be published in this journal (PNAS) at a later day.
▶ The article of EurekAlert! is here
Spontaneous Activity in Whisker-Innervating Region of Neonatal Mouse Trigeminal Ganglion
Iwasato Group / Laboratory of Mammalian Neural Circuits
Spontaneous Activity in Whisker-Innervating Region of Neonatal Mouse Trigeminal Ganglion.
P. Banerjee, F. Kubo, H. Nakaoka, R. Ajima, T. Sato, T. Hirata, T. Iwasato.
Scientific Reports (2022) 12, 16311 DOI:10.1038/s41598-022-20068-z
Spontaneous activity is the activity that occurs without any external sensory input. During the early postnatal period in mammals, such activity is thought to be crucial for the establishment of mature neural circuits. It remains unclear if the peripheral structure of the developing somatosensory system exhibits spontaneous activity, similar to that observed in the retina and cochlea of developing visual and auditory systems, respectively. By establishing an ex vivo calcium imaging system, here we discovered that neurons in the whisker-innervating region of the trigeminal ganglion (TG) of neonatal mice generate spontaneous activity. A small percentage of neurons showed some obvious correlated activity, and these neurons were mostly located close to one another. TG spontaneous activity was majorly exhibited by medium-to-large diameter neurons, a characteristic of mechanosensory neurons, and was blocked by chelation of extracellular calcium. Moreover, this activity was diminished by the adult stage. Spontaneous activity in the TG during the first postnatal week could be a source of spontaneous activity observed in the neonatal mouse barrel cortex.
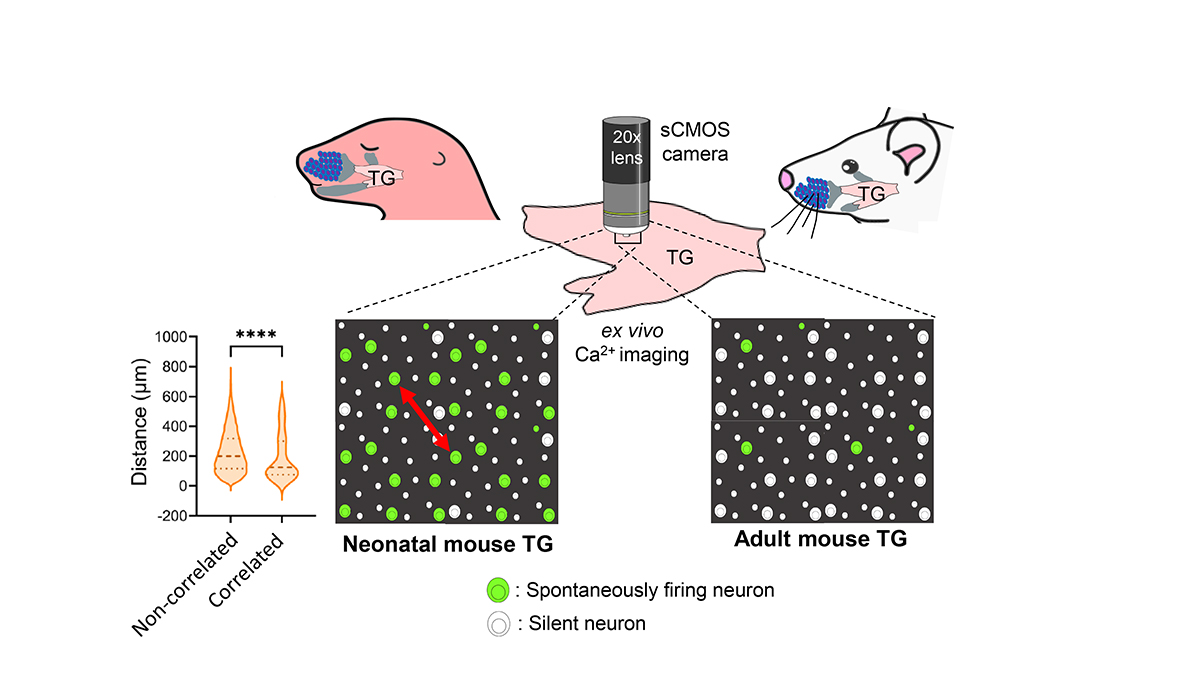
Sensory neurons of neonatal mouse trigeminal ganglion (TG) exhibit spontaneous activity ex vivo, which majorly diminishes by adult stage. Violin plot: Correlated neurons are closer to one-another, compared to non-correlated ones (Red arrow: Distance between two neurons).
Chromatin organization and DNA damage
Maeshima Group / Genome Dynamics Laboratory
Chromatin organization and DNA damage.
K. Minami, S. Iida, K. Maeshima
The Enzymes “DNA Damage and Double Strand Breaks” 2022 September 27 DOI:10.1016/bs.enz.2022.08.003
Free link (until 2022.11.16)
Genomic DNA is organized three-dimensionally in the nucleus as chromatin. Recent accumulating evidence has demonstrated that chromatin organizes into numerous dynamic domains in higher eukaryotic cells, which act as functional units of the genome. However, the cellular genome is constantly threatened by many sources of DNA damage (e.g., radiation). How do cells maintain their genome integrity when subjected to DNA damage?
In this review, we discuss how the compact state of chromatin safeguards the genome from radiation damage and chemical attacks. Together with recent genomics data, our finding (Takata et al. “Chromatin compaction protects genomic DNA from radiation damage”. PLOS ONE (2013). DOI: 10.1371/journal.pone.0075622) suggests that DNA compaction, such as chromatin domain formation, plays a critical role in maintaining genome integrity. But does the formation of such domains limit DNA accessibility inside the domain and hinder the recruitment of repair machinery to the damaged site(s) during DNA repair? To approach this issue, we first describe a sensitive imaging method to detect changes in chromatin states in living cells (single-nucleosome imaging). We then use this method to explain how cells can overcome potential recruiting difficulties; cells can decompact chromatin domains following DNA damage and temporarily increase chromatin motion (∼ DNA accessibility) to perform efficient DNA repair (Iida et al. “Single-nucleosome imaging reveals steady-state motion of interphase chromatin in living human cells” Science Advances (2022). DOI: 10.1126/sciadv.abn5626).
This review article will be published as the 3rd chapter in the book “The Enzymes vol.51: DNA Damage and Double Strand Breaks (Elsevier, 2022)”. This work was supported by JSPS and MEXT KAKENHI grants (19H05273 and 20H05936 to K. Maeshima), the Takeda Science Foundation (to K. Maeshima), and the Uehara Memorial Foundation (to K. Maeshima). K. Minami and S. Iida are SOKENDAI Special Researchers supported by JST SPRING JPMJSP2104.
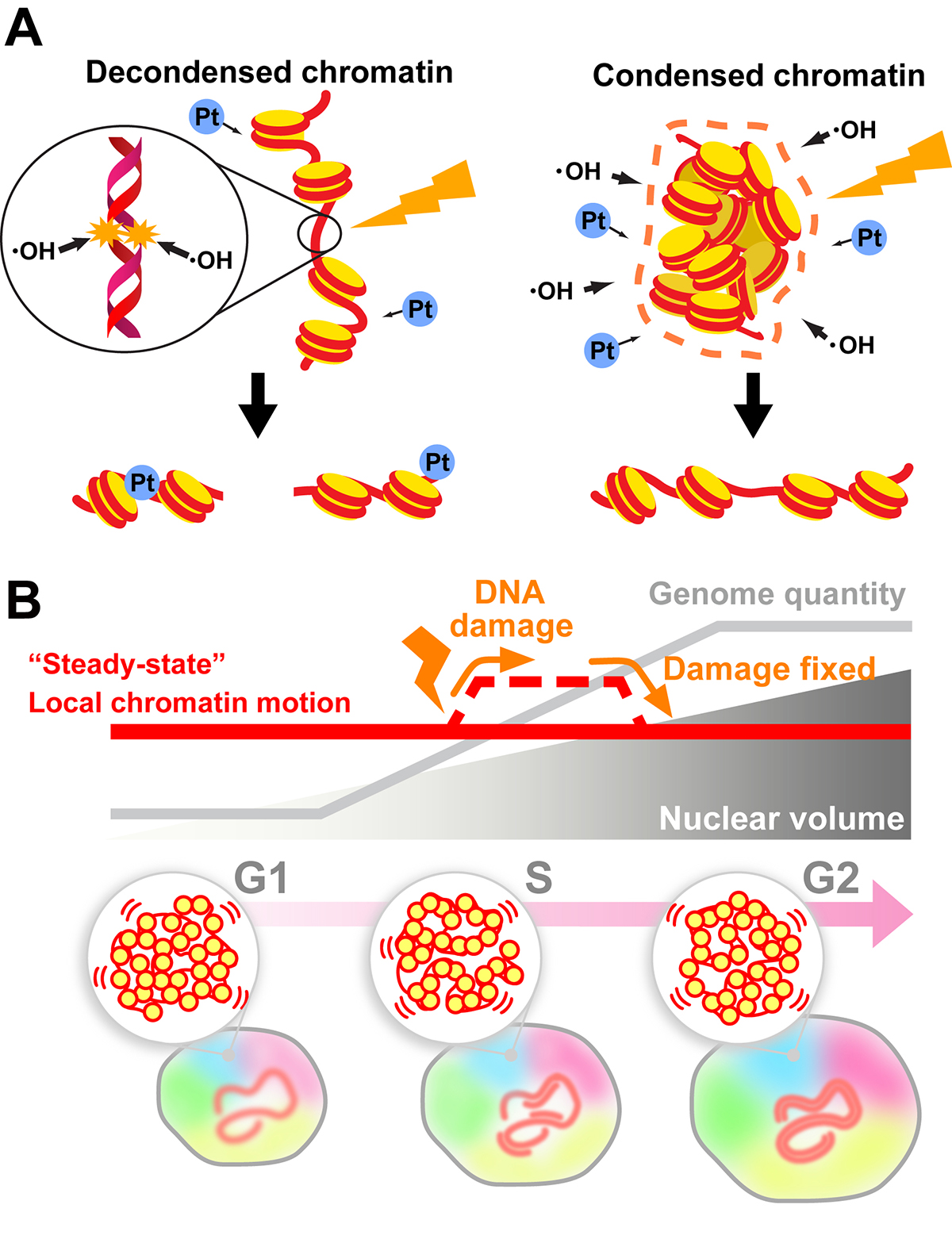
(B) Local chromatin motion persisted throughout interphase of the cell cycle (“steady-state” local chromatin motion; red line) can be transiently increased by a DNA damage response (dashed line). This temporal change in chromatin motion presumably occurs to facilitate access for DNA repair machinery.















