Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
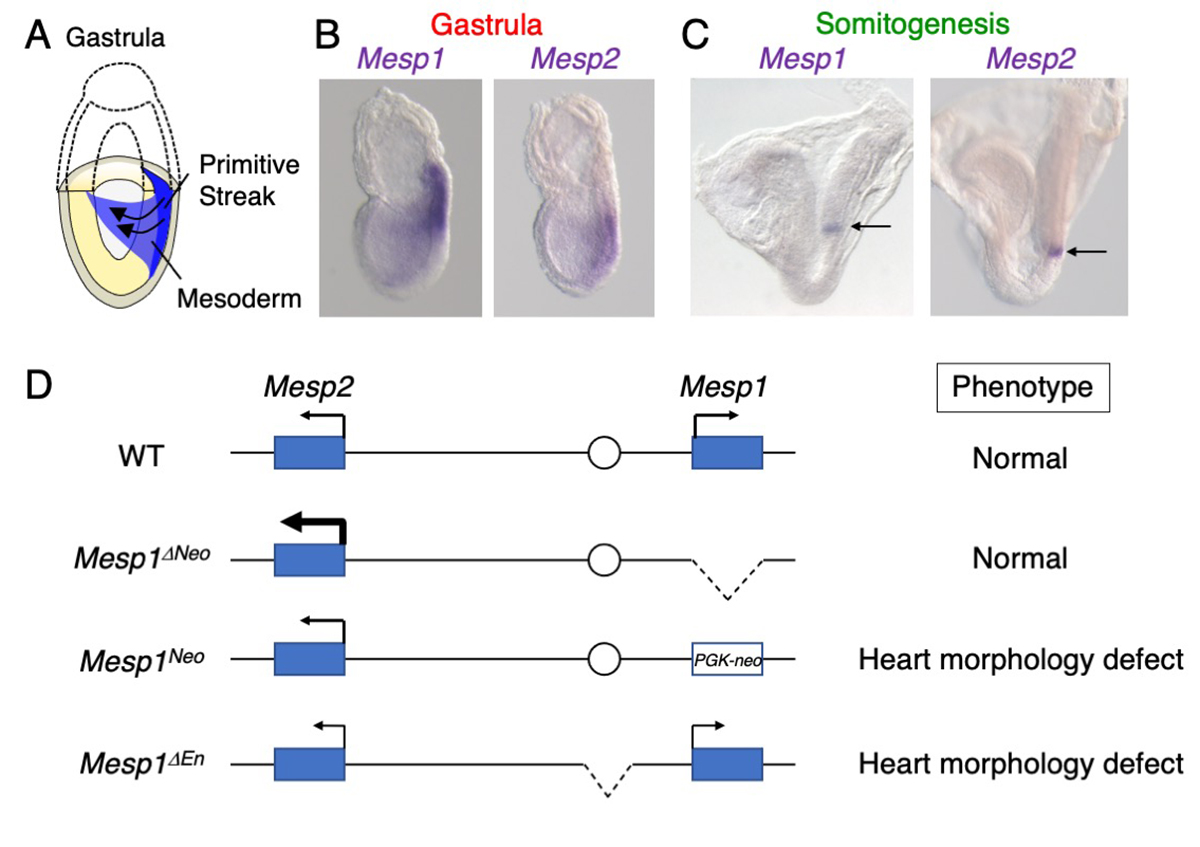
Mesoderm specification is determined by dose of MESP1 and MESP2.
Saga Group / Mammalian Development Laboratory
Formal proof of the requirement of MESP1 and MESP2 in mesoderm specification and their transcriptional control via specific enhancers in mice
Rieko Ajima, Yuko Sakakibara, Noriko Sakurai-Yamatani, Masafumi Muraoka and Yumiko Saga
Development (2021) 148, dev194613 DOI:10.1242/dev.194613
The mesodermal cells are derived from the primitive streak during gastrulation in mouse development (Figure A). These mesodermal cells give rise to the heart, somites, kidney, and limbs. MESP1 and MESP2 are transcriptional factors expressed in early mesoderm and boundary of newly forming somites (Figure B, C). The Mesp1/Mesp2 dKO embryos exhibited markedly severe mesoderm formation defects. However, MESP1 and MESP2 were thought to regulate distinct targets, because Mesp1 and Mesp2 single KO embryos display distinct phenotypes, heart morphogenesis defects and somite formation defects, respectively.
In this study, we established the Mesp1/Mesp2 mutants using genome editing techniques, and found the Mesp1/Mesp2 dKO embryos exhibited defects similar to the original Mesp1/Mesp2 dKO embryos. However, Mesp1 KO did not display any phenotypes. We noted up-regulation of Mesp2 in the Mesp1 KO embryos, suggesting that MESP2 rescues the loss of MESP1 in mesoderm specification. We also found that Mesp1 and Mesp2 expression in the early mesoderm is regulated by the common enhancer. Deletion of the enhancer caused the down-regulation of both genes, resulting in heart formation defects. This study suggests dosage-dependent roles of MESP1 and MESP2 in early mesoderm formation.
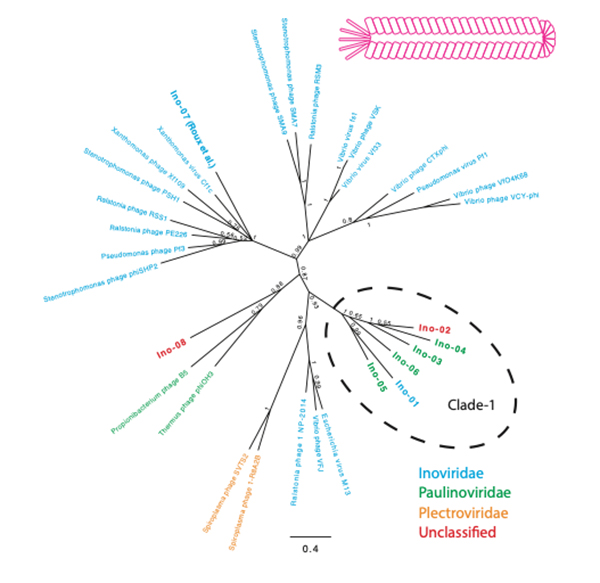
Comprehensive discovery of CRISPR-targeted terminally redundant sequences in the human gut metagenome: viruses, plasmids, and more
Press release
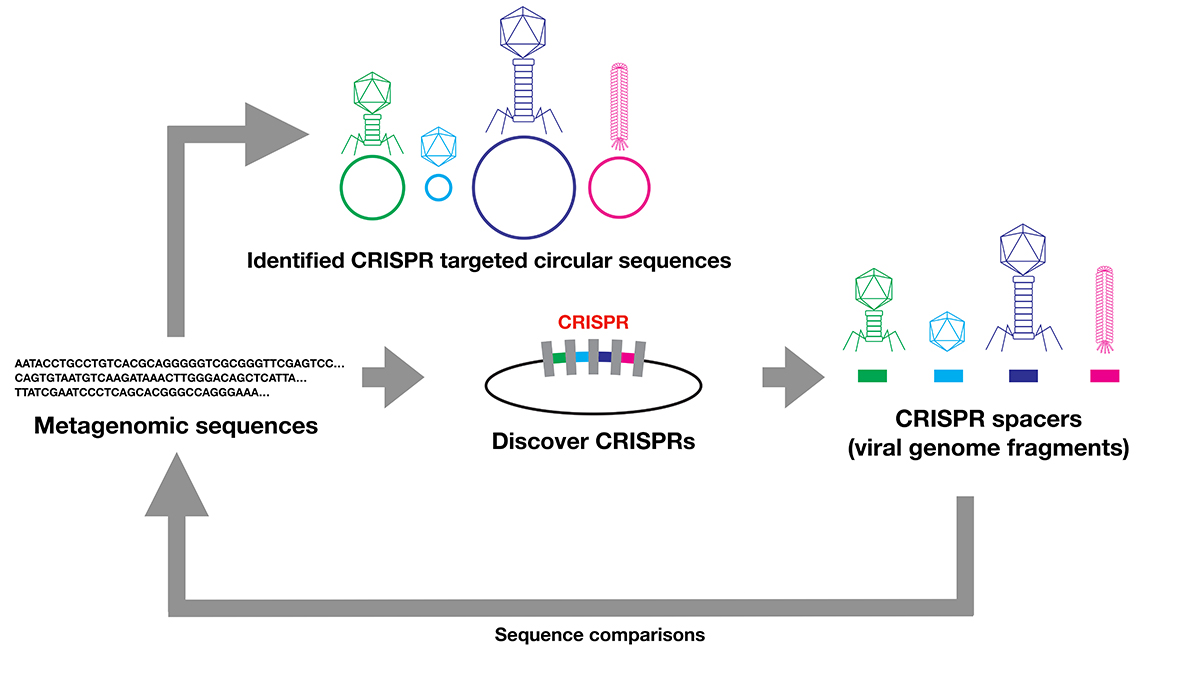
Comprehensive discovery of CRISPR-targeted terminally redundant sequences in the human gut metagenome: viruses, plasmids, and more
R. Sugimoto, L. Nishimura, P. T. Nguyen, J. Ito, N. F. Parrish, H. Mori, K. Kurokawa, H. Nakaoka, I. Inoue
PLOS Computational Biology (2021) 17, e1009428 DOI:10.1371/journal.pcbi.1009428
![]() Press release (In Japanese only)
Press release (In Japanese only)
The evolution and origins of viruses are long-standing questions in the field of biology. Viral genomes provide fundamental information to infer the evolution and origin of viruses. However, viruses are extraordinarily diverse, and there are no single genes shared across entire species. Several methods were developed to collect viral genomes from metagenome. To infer viral genomes from metagenome, previous approaches relied on reference viral genomes. We thought that such reference-based methods may not be sufficient to uncover diverse viral genomes; therefore, we developed a pipeline that utilizes CRISPR, a prokaryotic adaptive immunological memory. Using this pipeline, we discovered more than 10,000 positively complete CRISPR-targeted genomes from human gut metagenome datasets. A substantial portion of the discovered genomes encoded various types of capsid proteins, supporting the contention that these sequences are viral. Although the majority of these capsid-protein-coding sequences were previously characterized, we notably discovered Inoviridae genomes that were previously difficult to infer as being viral. Furthermore, some of the remaining unclassified sequences without a detectable capsid-protein-encoding gene had a notably low protein-coding ratio. Overall, our pipeline successfully discovered viruses and previously uncharacterized presumably mobile genetic elements targeted by CRISPR.
Source: R. Sugimoto et al., PLOS Computational Biology DOI:10.1371/journal.pcbi.1009428
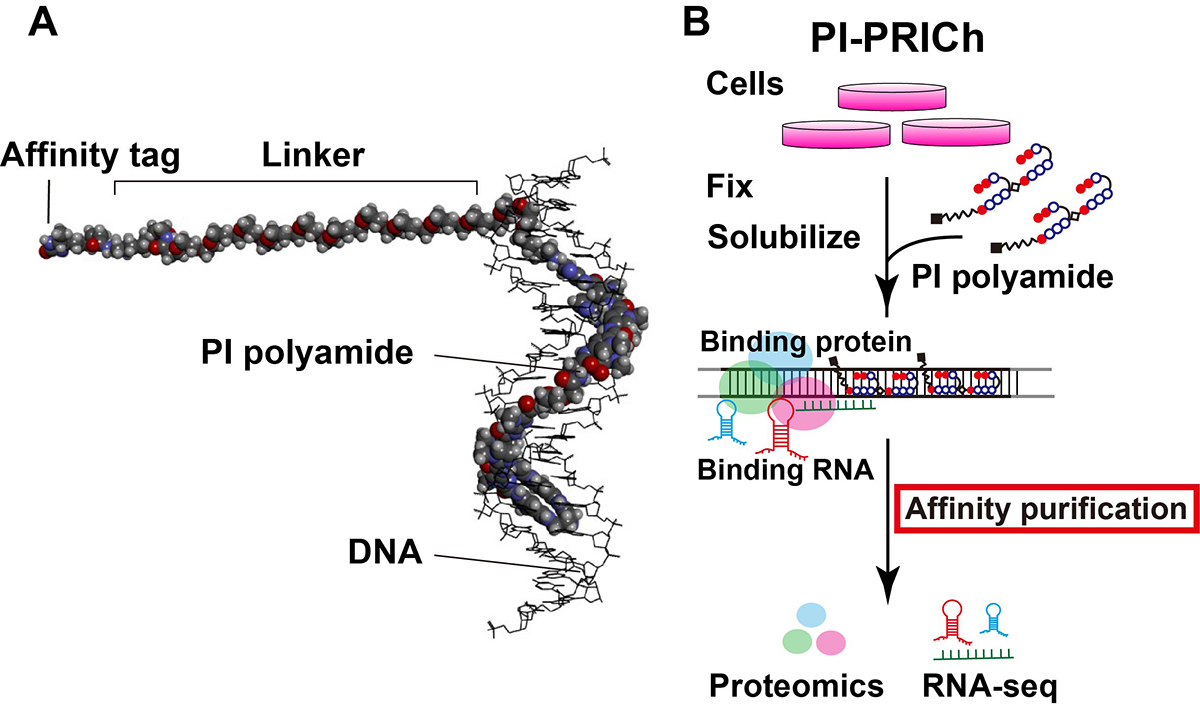
Telomere-specific chromatin capture using a pyrrole–imidazole polyamide probe
Maeshima Group / Genome Dynamics Laboratory
Telomere-specific chromatin capture using a pyrrole–imidazole polyamide probe for the identification of proteins and non-coding RNAs
Satoru Ide#*, Asuka Sasaki#, Yusuke Kawamoto, Toshikazu Bando, Hiroshi Sugiyama, Kazuhiro Maeshima
#Equally contributed, *Corresponding author
Epigenetics & Chromatin (2021) 14, 46 DOI:10.1186/s13072-021-00421-8
Background: Knowing chromatin components at a DNA regulatory element at any given time is essential for understanding how the element works during cellular proliferation, differentiation and development. A region-specific chromatin purification is an invaluable approach to dissecting the comprehensive chromatin composition at a particular region. Several methods (e.g., PICh, enChIP, CAPTURE and CLASP) have been developed for isolating and analyzing chromatin components. However, all of them have some shortcomings in identifying non-coding RNA associated with DNA regulatory elements.
Results: We have developed a new approach for affinity purification of specific chromatin segments employing an N-methyl pyrrole (P)−N-methylimidazole (I) (PI) polyamide probe, which binds to a specific sequence in double-stranded DNA via Watson–Crick base pairing as a minor groove binder (Figure 1A). This new technique is called proteomics and RNA-omics of isolated chromatin segments (PI-PRICh). Using PI-PRICh to isolate mouse and human telomeric components, we found enrichments of shelterin proteins, the well-known telomerase RNA component (TERC) and telomeric repeat-containing RNA (TERRA) When PI-PRICh was performed for alternative lengthening of telomere (ALT) cells with highly recombinogenic telomeres, in addition to the conventional telomeric chromatin, we obtained chromatin regions containing telomeric repeat insertions scattered in the genome and their associated RNAs.
Conclusion: PI-PRICh reproducibly identified both the protein and RNA components of telomeric chromatin when targeting telomere repeats. PI polyamide is a promising alternative to simultaneously isolate associated proteins and RNAs of sequence-specific chromatin regions under native conditions, allowing better understanding of chromatin organization and functions within the cell (Figure 1B).
This work was supported by an NIG-JOINT (2015-B6), JSPS grants (JP17J10836 to A.S.; 15H01361 and 21H02535 to S.I.; 20H05936 and 21H02453 to K.M.), the Takeda Science Foundation to K.M. and the Uehara Memorial Foundation to K.M.. A.S. was a JSPS Fellow (DC2).
A sugar utilization phenotype contributed to the formation of genetic exchange communities in the ecological niches among lactic acid bacteria.
Arita Group / Biological Networks Laboratory
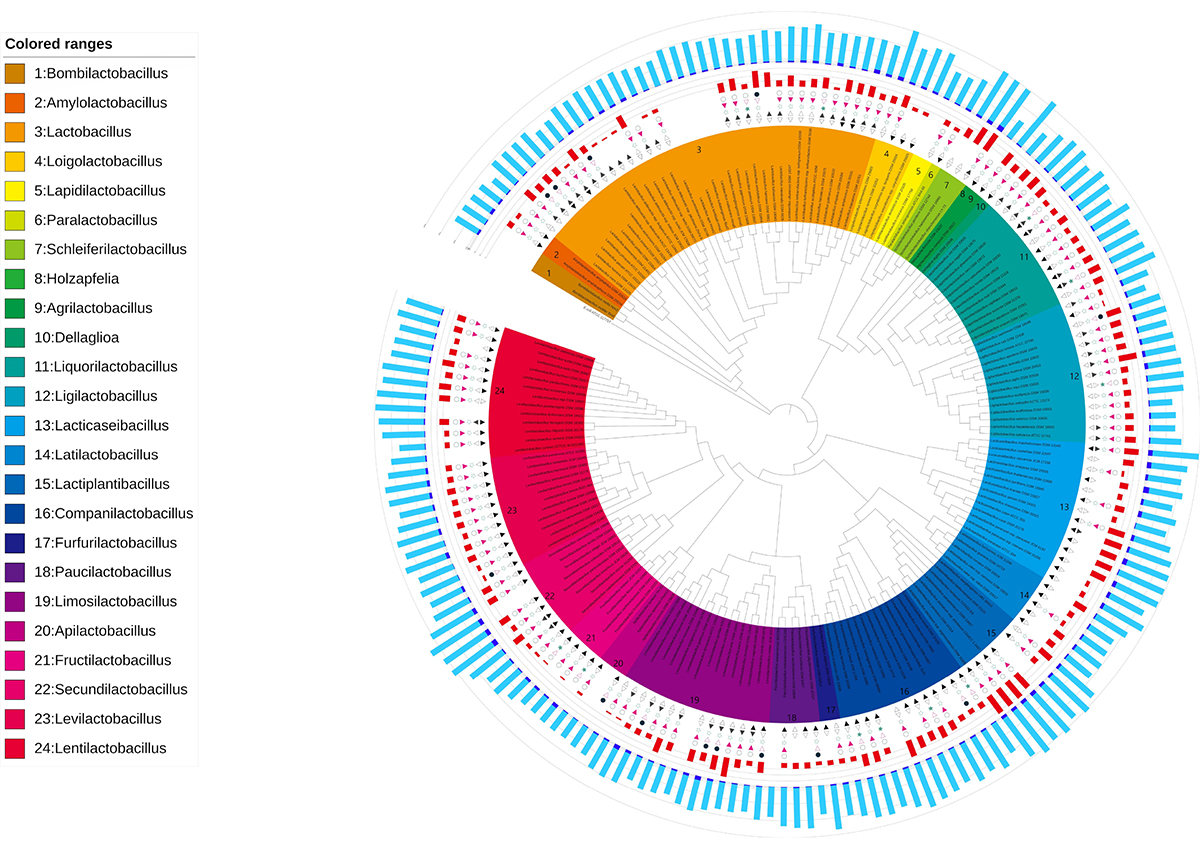
A sugar utilization phenotype contributes to the formation of genetic exchange communities in lactic acid bacteria
Shinkuro Takenaka, Takeshi Kawashima, Masanori Arita.
FEMS Microbiology Letters (2021) 368, fnab117 DOI:10.1093/femsle/fnab117
In prokaryotes, a major contributor to genomic evolution is the exchange of genes via horizontal gene transfer (HGT). Areas with a high density of HGT networks are defined as genetic exchange communities (GECs). Although some phenotypes associated with specific ecological niches are linked to GECs, little is known about the phenotypic influences on HGT in bacterial groups within a taxonomic family. Thanks to the published genome sequences and phenotype data of lactic acid bacteria (LAB), it is now possible to obtain more detailed information about the phenotypes that affect GECs. Here, we have investigated the relationship between HGT and internal and external environmental factors for 178 strains from 24 genera in the Lactobacillaceae family. We found a significant correlation between strains with high utilization of sugars and HGT bias. The result suggests that the phenotype of the utilization of a variety of sugars is key to the construction of GECs in this family. This feature is consistent with the fact that the Lactobacillaceae family contributes to the production of a wide variety of fermented foods by sharing niches such as those in vegetables, dairy products and brewing-related environments. This result provides the first evidence that phenotypes associated with ecological niches contribute to form GECs in the LAB family.
Source: S. Takenaka, et al., DOI: 10.1093/femsle/fnab117
Figure at the beginning: Phylogenetic tree based on the 16S rRNA genes of the LAB strains with the phenotypic and genomic features identified. The inner band shows species colored by genus. The next five symbols show phenotypic characteristics for each LAB strain; first inward-facing triangle indicates the growth at 15°C, second outward-facing triangle indicates the growth at 45°C, third star indicates the micro aerophilic, fourth red inward-facing indicates facultatively anaerobic and fifth circle indicates obligate anaerobic. A filled symbol means the strain has the phenotype, and an open symbol means that it does not. A blank means that there is no relevant information available. The next red band shows the number of sugar types that can be utilized. The outer bands show the number of coding sequences (CDS) for each strain: navy blue indicates the estimated number of CDS acquired by the horizontal gene transfer (HGT) and light blue indicates the number of native CDS.
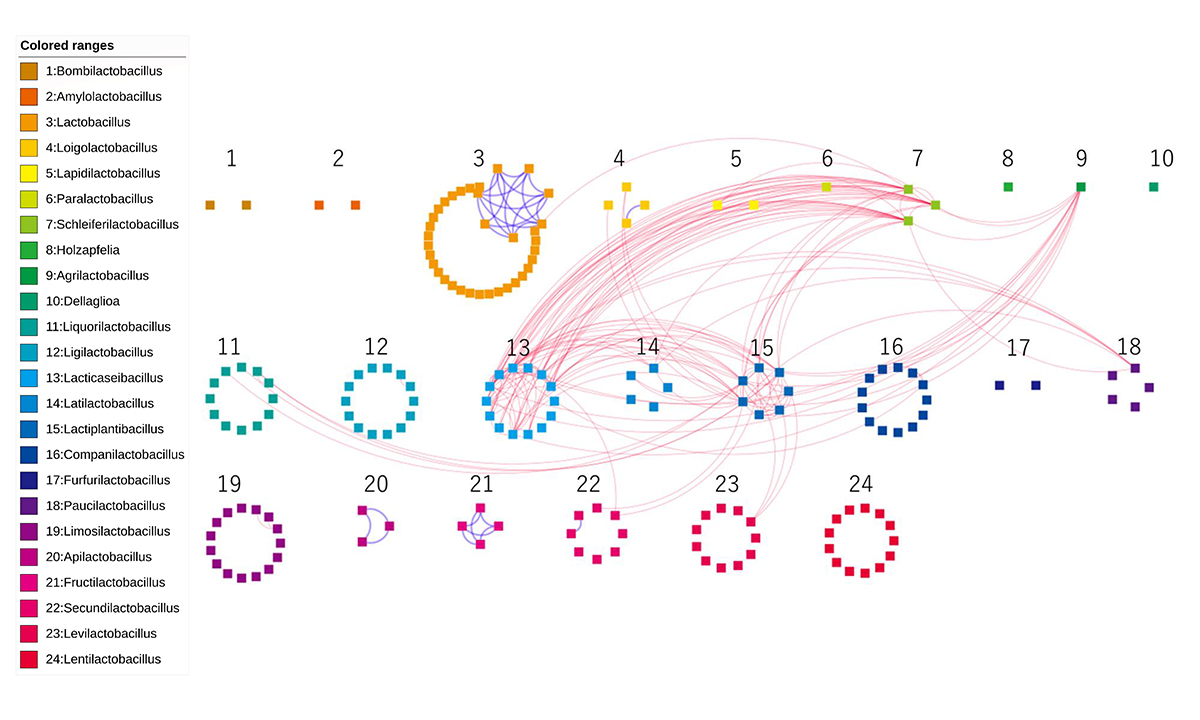
Figure: The networks for the generalist and specialist group orthologs. Each of the 178 nodes represents an LAB genome, which are colored and numbered by genus. Edges of dotted-red/solid-blue were created between two genomes when the number of sharing generalist/specialist group orthologs was more than five.
New faculty appointment as of October 1st
Dr. Keisuke Yonehara joined NIG as a professor on October 1.
YONEHARA, Keisuke : Multiscale Sensory Structure Laboratory

- YONEHARA, Keisuke professor















