Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Liquid-like chromatin in the cell and the cohesin activity
Maeshima Group / Genome Dynamics Laboratory
Liquid-like chromatin in the cell: What can we learn from imaging and computational modeling?
Yuji Itoh, Esmae J. Woods, Katsuhiko Minami, Kazuhiro Maeshima, and Rosana Collepardo-Guevara
Current Opinion in Structural Biology 71, 123-135 (2021) DOI:10.1016/j.sbi.2021.06.004
The loopy world of cohesin.
Kazuhiro Maeshima and Shiori Iida
eLife 10, e71585 (2021) DOI:10.7554/eLife.71585
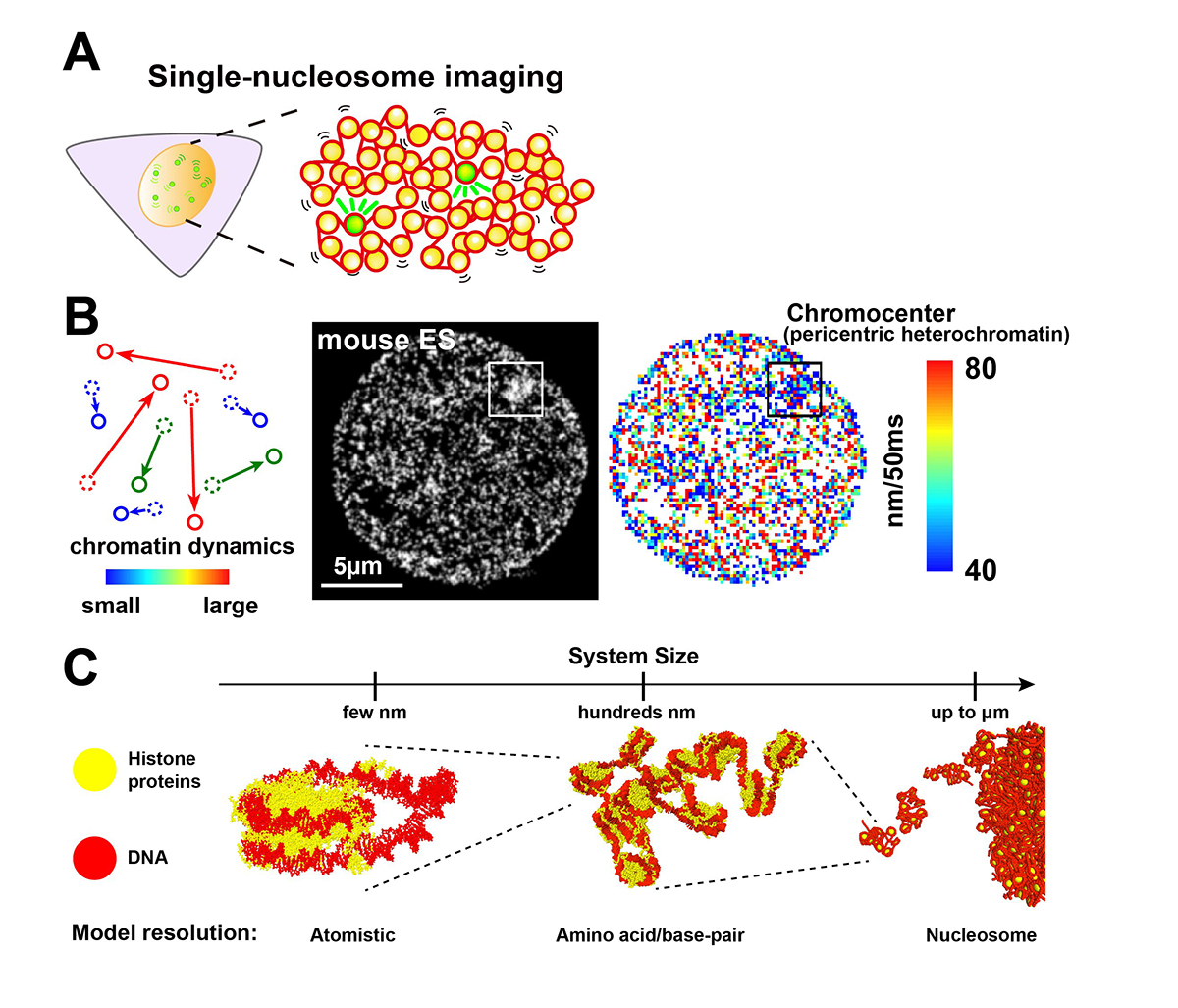
Chromatin in eukaryotic cells is a negatively charged long polymer consisting of DNA, histones, and various associated proteins. With its highly charged and heterogeneous nature, chromatin structure varies greatly depending on various factors (e.g., chemical modifications and protein enrichment) and the surrounding environment (e.g., cations): From a 10-nm fiber, a folded 30-nm fiber, to chromatin condensates/droplets. Recent advanced imaging such as single-nucleosome imaging (Figure 1A) has observed that chromatin exhibits a dynamic liquid-like behavior and undergoes structural variations within the cell (Figure 1B). Current computational modeling has made it possible to reconstruct the liquid-like chromatin in the cell by dealing with a number of nucleosomes on multi-scale levels, and has become a powerful technique to inspect the molecular mechanisms giving rise to the observed behavior, which imaging methods cannot do on their own (Figure 1C). Based on new findings from both imaging and modeling studies, we discuss the dynamic aspect of chromatin in living cells and its functional relevance.
This work was supported by JSPS grant (19K23735, 20J00572, 20H05936, 21H02453), the Takeda Science Foundation, the Uehara Memorial Foundation, NIG Postdoctoral Fellowship, JSPS Postdoctoral Fellowship (PD).
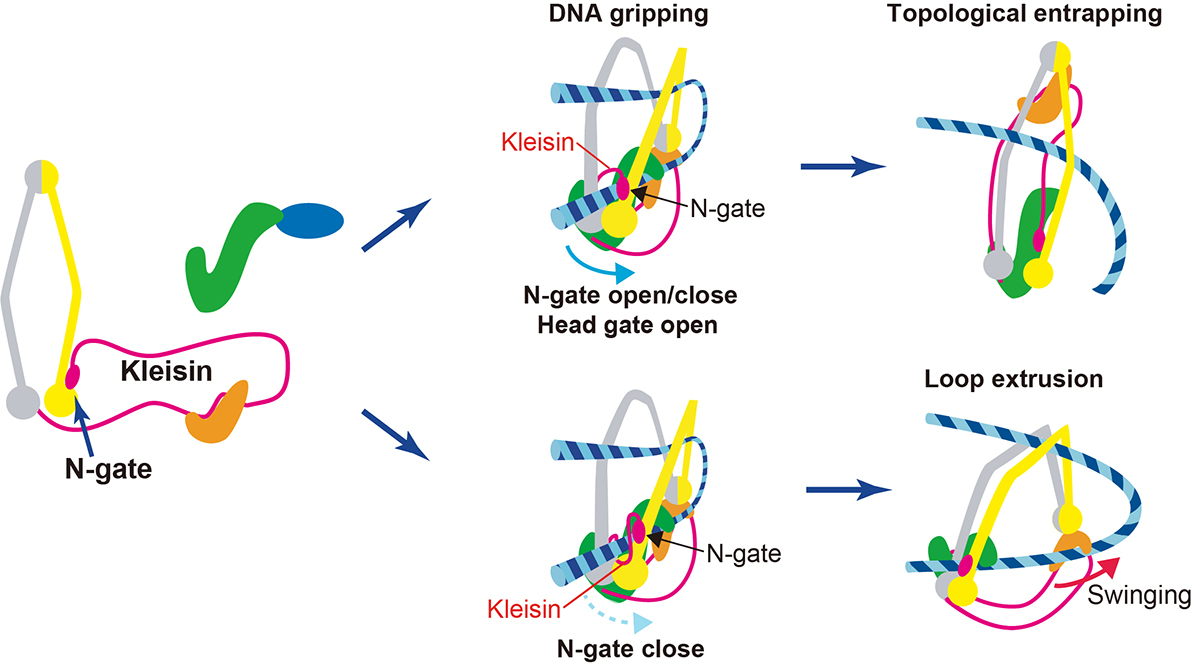
Also, on July 26th, a professor at Genome Dynamics Laboratory Kazuhiro Maeshima and SOKENDAI Ph.D. student Shiori Iida published an Insight paper in eLife. Chromatin higher-order structures, such as chromatin loop domains, are critical for chromatin to perform various functions in the cell. The formation of these chromatin loops is thought to be mediated by a ring-shaped molecular complex, cohesin (Figure 2, left). Currently, the mechanism of chromatin loop formation is a hot topic in cell biology, and a model called loop extrusion, in which cohesin pushes DNA out of the ring, has been attracting much attention. In fact, cohesin has been shown to extrude naked DNA in vitro. Recently, Dr. Frank Uhlmann and his colleagues have shown how cohesin excludes DNA loops in vitro (Figure 2, bottom)(Higashi et al., “A Brownian ratchet model for DNA loop extrusion by the cohesin complex”. eLife, 2021 DOI:10.7554/eLife.67530). Based on the Higashi et al. paper, Maeshima and Iida discussed the intracellular behavior of cohesin and whether the loop extrusion reported really occurs in the cell.
Estrogens enhance female-specific itch
Press release

Estrogens influence female itch sensitivity via the spinal gastrin-releasing peptide receptor neurons
K. Takanami*, D. Uta, K. Matsuda, M. Kawata, E. Carstens, T. Sakamoto, and H. Sakamoto
*Corresponding author
PNAS 118, e2103536118 (2021) DOI:10.1073/pnas.2103536118
![]() Press release (In Japanese only)
Press release (In Japanese only)
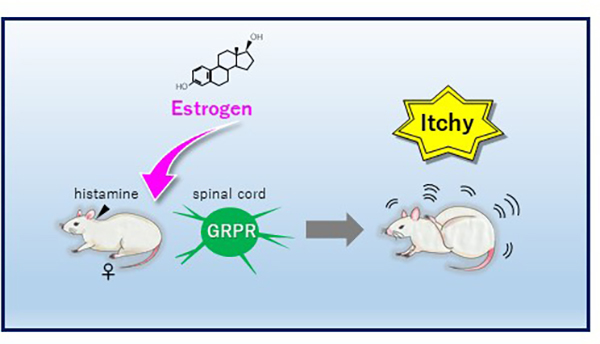
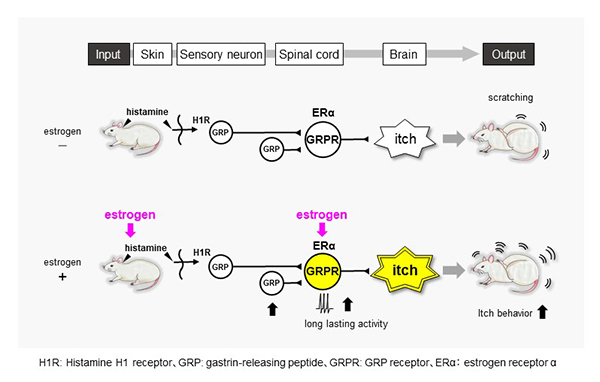
“Itch” sensitivity in women changes during periods when female sex hormones fluctuate, such as during pregnancy and menopause. Especially, many women exhibit itch symptoms during pregnancy, but the underlying mechanism of the change in itch sensitivity is unknown. Here Dr. Takanami et al. demonstrate that estradiol, but not progesterone, enhances histamine-evoked itch-related scratching behavior in female rats. This is associated with the enhancement of activity of “gastrin-releasing peptide receptor (GRPR)” neurons in the spinal cord (Fig. 1). The findings suggest that female hormone, estrogens selectively enhance histamine-evoked itching by regulating spinal GRP system in females (Fig. 2). This may account for why itch sensation varies across the female lifecycle and provides a novel basis for treating itchy diseases in females.
▶ This article was seleceted “In This Issue” of PNAS.
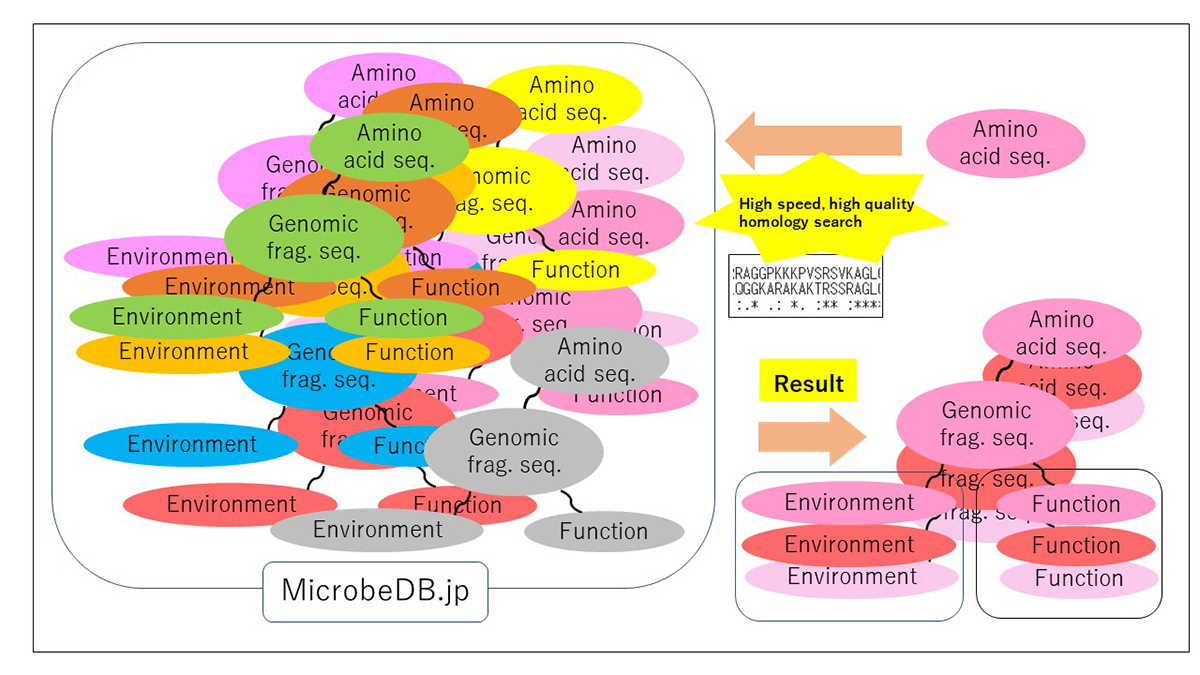
PZLAST: an ultra-fast amino acid sequence similarity search server against public metagenomes

PZLAST: an ultra-fast amino acid sequence similarity search server against public metagenomes
H. Mori, H. Ishikawa, K. Higashi, Y. Kato, T. Ebisuzaki, K. Kurokawa
Bioinformatics 2021 July 7 DOI:10.1093/bioinformatics/btab492
![]() Press release (In Japanese only)
Press release (In Japanese only)
Similarity searches of amino acid sequences against the public metagenomic data can provide users insights about the function of sequences based on the environmental distribution of similar sequences. However, a considerable reduction in the amount of data or the accuracy of the result was necessary to conduct sequence similarity searches against public metagenomic data, because of the vast data size more than Terabytes. Here, we present an ultra-fast service for the highly accurate amino acid sequence similarity search, called PZLAST, which can search the user’s amino acid sequences to several Terabytes of public metagenomic sequences in approximately 10-20 minutes. PZLAST accomplishes its search speed by using PEZY-SC2, which is a MIMD many-core processor. Results of PZLAST are summarized by the ontology-based environmental distribution of similar sequences. PZLAST can be used to predict the function of sequences and mine for homologs of functionally important gene sequences.
Source: H. Mori, et al., Bioinformatics DOI: 10.1093/bioinformatics/btab492
▶ PZLAST is available here.