Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Glia-neuron interactions underlie state transitions to generalized seizures
Press release
Glia-neuron interactions underlie state transitions to generalized seizures
Carmen Diaz-Verdugo, Sverre Myren-Svelstad, Ecem Aydin, Evelien van Hoeymissen, Celine Deneubourg, Silke Vanderhaeghe, Julie Vancraeynest, Robbrecht Pelgrims, Mehmet LLyas Cosacak, Akira Muto, Caghan Kizil, Koichi Kawakami6, Nathalie Jurisch-Yaksi & Emre Yaksi
Nature Communications 10, Article number: 3830 (2019) DOI:10.1038/s41467-019-11739-z
Press release (In Japanese only)
Brain activity and connectivity alters drastically during the generalization of epileptic seizures. Throughout this transition, brain networks shift from a balanced resting state to a hyperactive and hypersynchronous state, spreading across the brain. It is however less clear which mechanisms underlie these state transitions. By studying neuronal and glia activity across the zebrafish brain, we observed striking differences between these networks. During pre-ictal period, neurons displayed a small increase in synchronous activity only locally, while the entire glial network was highly active and strongly synchronized across large distances. We observed that the transition from a pre-ictal state to a generalized seizure leads to an abrupt increase in neural activity and connectivity, which is accompanied by a strong functional coupling between glial and neuronal networks. Optogenetic activation of glia induced strong and transient burst of neuronal activity, emphasizing a potential role for glia-neuron connections in the generalization of epileptic seizures.
Sourse: Nature Communications 10, Article number: 3830 (2019)
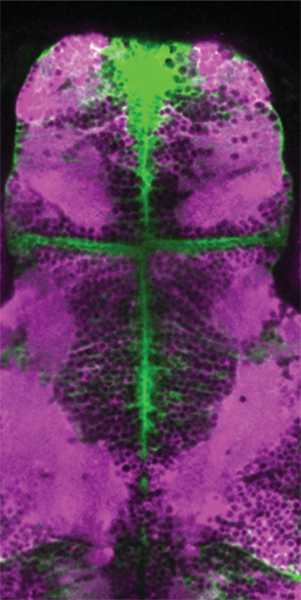
Fig: An image of the brain of the GFAP:Gal4;UAS:GCaMP6s;jRCaMP1a fish.
▶This study is based on the previous study.
The making of ‘Fancy Mouse’: Study reveals true cause of colorful hair on popular East Asian pet mice
Press release
Nested retrotransposition in the East Asian mouse genome causes the classical nonagouti mutation
Akira Tanave, Yuji Imai, Tsuyoshi Koide
Communications Biology 2 August 2019 10.1038/s42003-019-0539-7
EurekAlert! link about this artcle
For the past few hundred years, the colorful hair and unique patterns of the so-called “Fancy Mouse” have made them the stars of pet shows in Japan and beyond. Now, scientists have finally revealed the true cause of the genetic mutation responsible for the iconic black pigmentation in the popular East Asian pet.
Their findings were published on August 2, 2019 in Communications Biology. read more>

Fig: The mutated colors in Japanese fancy mice, JF1, gave them a Giant Panda look. The C57BL/6 (B6) strain has black coat color, nonagouti. Japanese wild strain MSM has agouti coat color.
*Credit: Tsuyushi Koide, the National Institute of Genetics (NIG) in Japan
▶ You can read the deeper story behind the paper below.
Reexamining the a of Mouse Genetics A to Z
▶Associate Prof. Koide posted “Comment” about this article.
A role for the rare endogenous retrovirus β4 in development of
Japanese fancy mice(PDF)
DFAST: Prokaryotic genome annotation pipeline for data submission to DDBJ
DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication
Yasuhiro Tanizawa, Takatomo Fujisawa, Yasukazu Nakamura Bioinformatics 34(6) 1037-9, 2018 DOI:10.1093/bioinformatics/btx713Current technology of high throughput sequencing forces microbiologists to deal with more and more genomic data than before. In addition, annotated genome data must be deposited in the public sequence database; however, its complicated process is an another burden for researchers. We have developed an automatic prokaryotic genome annotation pipeline called DDBJ Fast Annotation and Submission Tool (DFAST) aiming to assist data submission to DDBJ. DFAST is available as a web tool, which allows users to create data submission files using graphical user interface, and also as a stand-alone tool, which is well suited for massive data processing on a local machine. DFAST can annotate a typical-sized bacterial genome within 5 minutes and has unique features such as pseudogene detection. It is available at https://dfast.ddbj.nig.ac.jp.
DFAST will facilitate the use of genomic data in microbial studies.
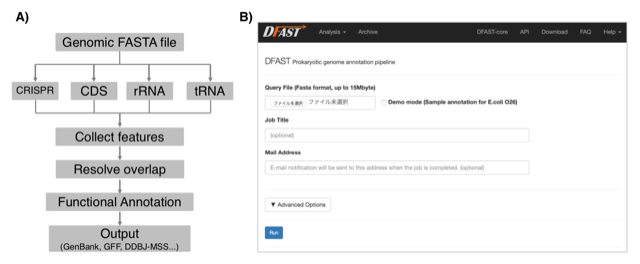
Figure: A) Annotation workflow of DFAST. B) Job submission form of the DFAST web server.
Visualization of autophagy in rice anther tapetum
Monitoring autophagy in rice tapetal cells during pollen maturation.
Shigeru Hanamata, Jumpei Sawada, Bunki Toh, Seijiro Ono, Kazunori Ogawa, Togo Fukunaga, Ken-Ichi Nonomura, Takamitsu Kurusu, Kazuyuki Kuchitsu.
Plant Biotechnology 36 (2): 99-105 (2019) DOI:10.5511/plantbiotechnology.19.0417a
Autophagy, a major catabolic pathway in eukaryotic cells, plays critical roles in the recycling of proteins and lipids, and is involved in many developmental processes. It plays important roles in pollen maturation and required for tapetal programmed cell death (PCD), postmeiotic anther development and nutrient supply to the pollens in rice (Kurusu et al. 2014). In this paper, we introduce an in vivo imaging technique to analyze the dynamics of autophagy in rice tapetum by expressing GFP-ATG8, a marker for autophagosomes under the control of tapetum-specific promoters. The results from imaging analyses including 3-dimensional visualization revealed that autophagy is dramatically induced at the specific stages throughout the tapetal cells just before their PCD process (Fig. 1). This monitoring system offers a powerful tool to analyze the regulation of autophagy in rice tapetum during anther development, and contribute to the research to increase the grain quality and yield of rice.
This work was supported in part by the NIG-Joint program (84A2018).
Glossary
Autophagosome: A spherical structure with double layer membranes. It is the key structure in macroautophagy, the intracellular degradation system for cytoplasmic contents.
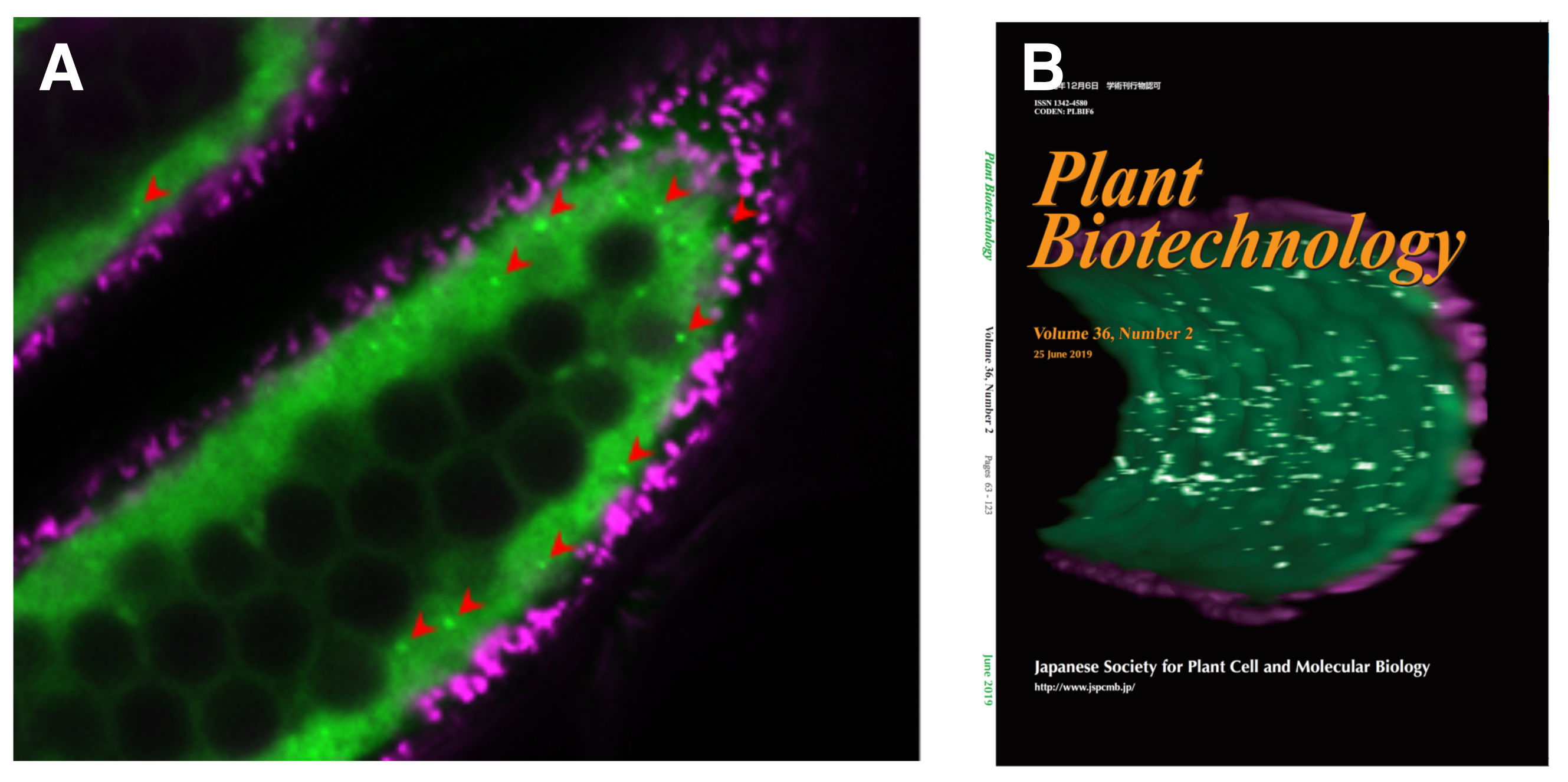
Figure: Visualization of the dynamics of autophagosomes/autophagy-related structures in rice anthers.
(A) A confocal fluorescence image of cross-section of rice anthers indicates GFP (green) and autofluorescence (magenta) of chlorophyll, respectively. Arrowheads indicate punctate signals of GFP-ATG8 (autophagosomes/autophagy-related structures). (B) Three-dimensional reconstruction of the tapetum undergoing autophagy.
Protein factors increasing yield of a biofuel precursor in microscopic algae
Press release
LIPID REMODELING REGULATOR 1 (LRL1) is differently involved in the phosphorus-depletion response from PSR1 in Chlamydomonas reinhardtii
Nur Akmalia Hidayati, Yui Yamada-Oshima, Iwai Masako, Takashi Yamano, Masataka Kajikawa, Nozomu Sakurai, Kunihiro Suda, Kanami Sesoko, Koichi Hori, Takeshi Obayashi, Mie Shimojima, Hideya Fukuzawa, Hiroyuki Ohta
The Plant Journal 27 July 2019 DOI:10.1111/tpj.14473
EurekAlert! link about this artcle
As an alternative to traditional fossil fuels, biofuels represent a more environmentally friendly and sustainable fuel source. Plant or animal fats can be converted to biofuels through a process called transesterification. In particular, the storage molecule triacylglycerol (TAG), found in microscopic algae, is one of the most promising sources of fat for biofuel production, as microalgae are small, easy to grow, and reproduce quickly. Therefore, increasing the yield of TAG from microalgae could improve biofuel production processes. With this ultimate goal in mind, Professor Hiroyuki Ohta from the Tokyo Institute of Technology and colleagues investigated the conditions under which the model microalga Chlamydomonas reinhardtii produces more TAG. read more>
This research was published in The Plant Journal at 27 July 2019.
You can get the tools and databases that was the basis for metabolome analysis in this research from the following site. KOMICS (The Kazusa Metabolomics Portal)
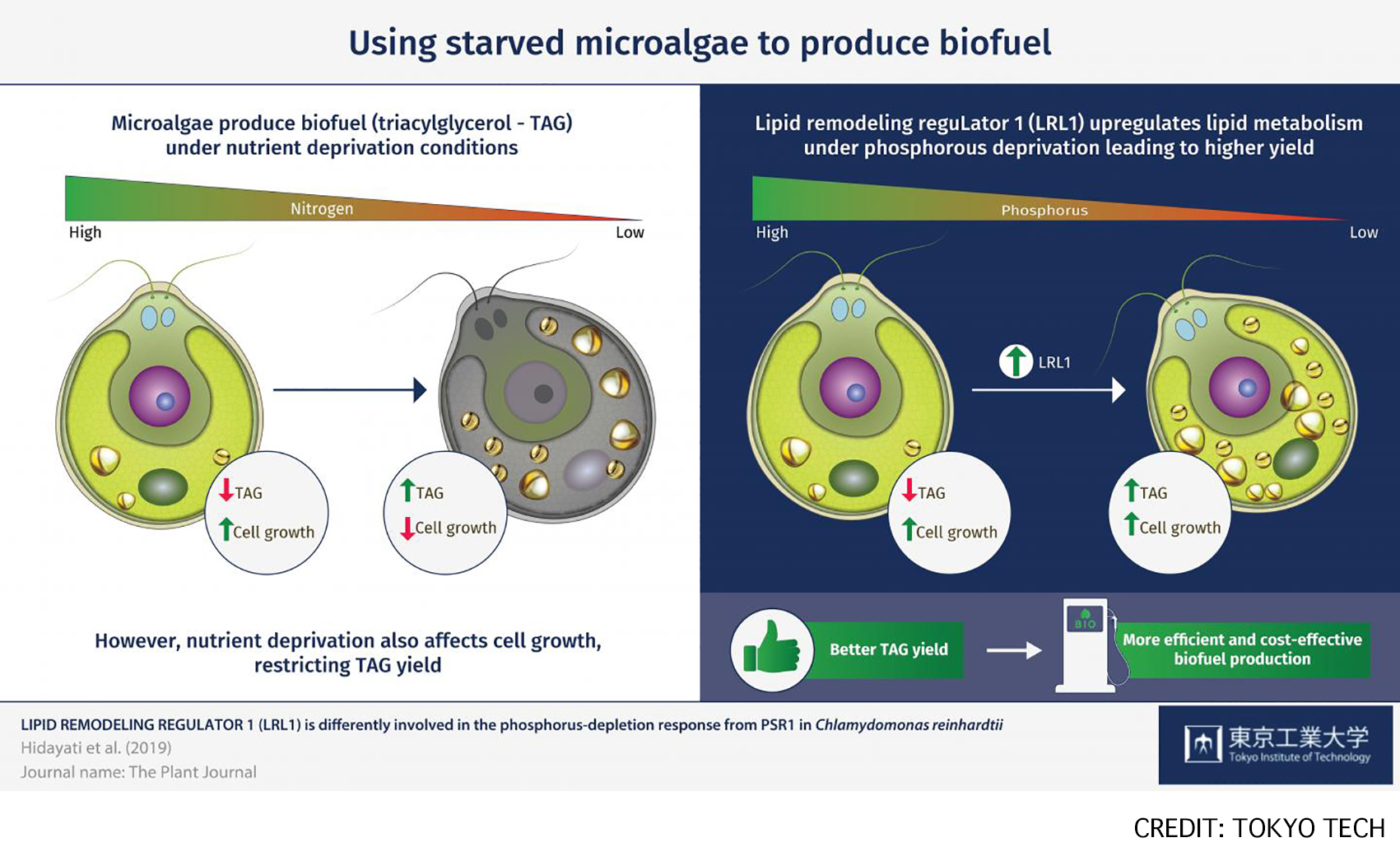
Fig: Microalgae are a promising source of biofuel feedstock, as they produce triacylglycerol (TAG) as a major storage lipid, especially under nutrient-depleted conditions. LRL1 was involved in the regulatory mechanism during the later stage of P-starvation in C. reinhardtii, as its regulation might depend on P-status, cell growth, and other factors.















