Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
A universal correlation between the density of the DNA and the length of mitotic chromosome
Cell Architecture Laboratory / Kimura Group
Scaling relationship between intra-nuclear DNA density and chromosomal condensation in metazoan and plant.
Hara Y, Adachi K, Kagohashi S, Yamagata K, Tanabe H, Kikuchi A, Okumura S-I, Kimura A.
Chromosome Science, 19, 43-49 (2016). DOI:10.11352/scr.19.43
Because the fundamental structure of chromosomes is conserved across eukaryotes, it might be assumed that an increase in the number of DNA base-pairs in a chromosome would lead to a corresponding increase in the physical length of chromosome. This does not appear to be the case, however. We compared the lengths of mitotic chromosome from several diverse species to determine the relationship between chromosome length, number of base-pairs, and the extent of chromosome packing. We found that all species share the same relationship among these, indicating that as base-pairs are added, chromosomes become more tightly packed so that the overall length increases less than expected. Our results suggest that instead of being related to the number of DNA base-pairs, chromosome length might be proportional to the surface area of the nucleus. This may be due to the need for the chromosomes to fit within a nuclear area known as the metaphase plate during mitosis, which occurs during cellular reproduction. This study provides insight into the features that drive the evolution of genome, chromosome, nucleus, and cell size and indicates that these characteristics are shared across eukaryotes.
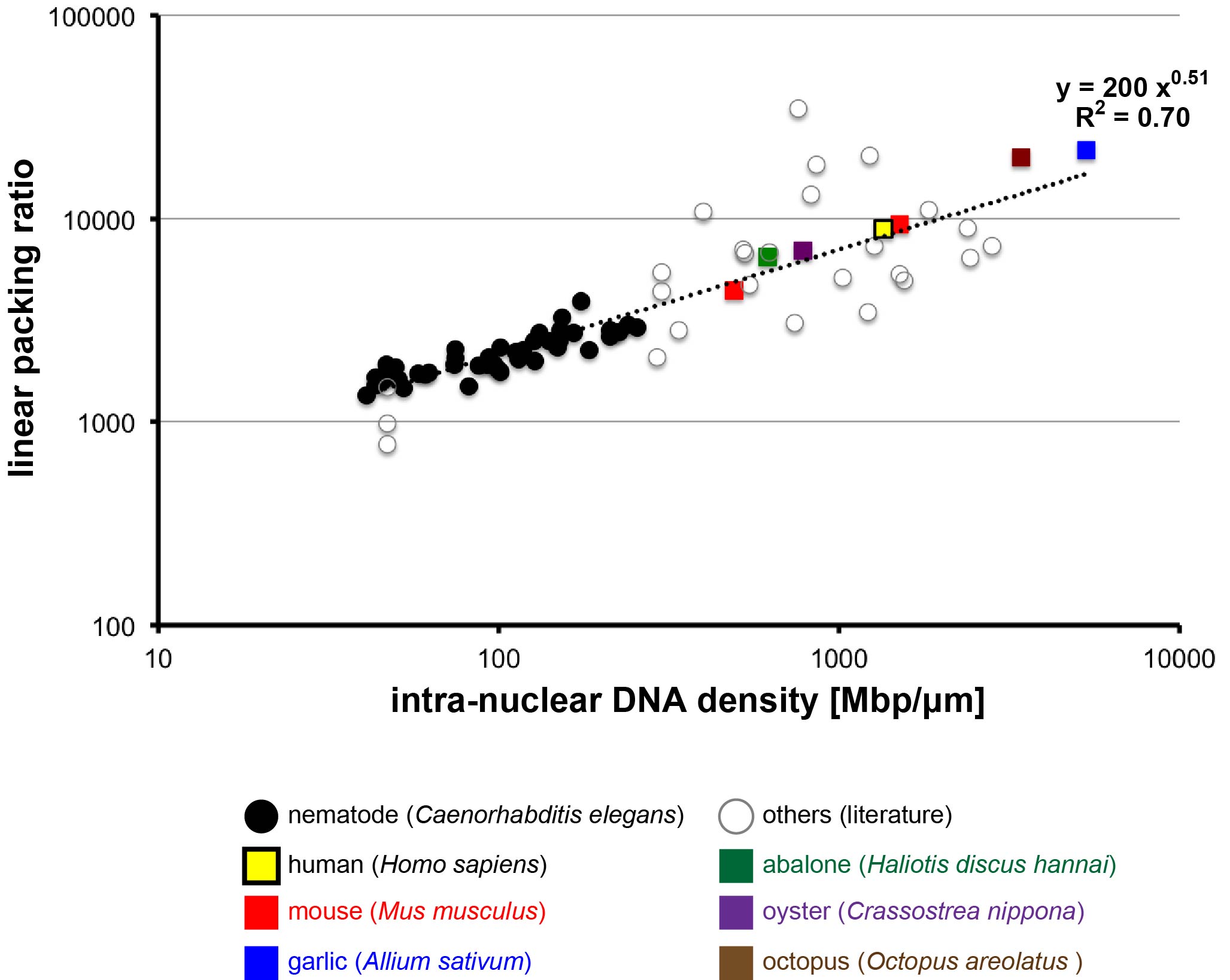
Figure. The degree of chromosome condensation (linear packing ratio) is correlated with intra-nuclear DNA density across species (Figure 1 of this paper). The double logarithmic plot of linear packing ratio against intra-nuclear DNA density is shown. The values obtained in this study are denoted with colored squares.
The timeline of chromosome organization in early embryogenesis
Cell Architecture Laboratory / Kimura Group
Reduction in chromosome mobility accompanies nuclear organization during early embryogenesis in Caenorhabditis elegans.
Arai R, Sugawara T, Sato Y, Minakuchi Y, Toyoda A, Nabeshima K, Kimura H, Kimura A.
Scientific Reports, 7, 3631 (2017). DOI:10.1038/s41598-017-03483-5
Many of us may imagine the DNA inside our cells as a jumble of noodles. However, DNA is more organized than this inside the nucleus, with chromosomes occupying distinct nuclear territories, for example. It is unclear though whether this organization is always present or whether it appears at some point during development after fertilization of the egg. We studied chromosome organization by observing the mobility of chromosomes inside in the nucleus in developing nematode embryos from the 2-cell to the 48-cell stage. We found that chromosome mobility decreases in 8-cell embryos, suggesting the initiation of chromosome organization at this point. Chromosome organization in the nucleus is important for gene expression and may have other purposes as well. For example, we found that in nematodes, the timing of chromosome organization coincides with the appearance of epigenetic marks, which regulate gene expression, and of a nuclear domain called the nucleolus. Now that we have identified the timeline of nuclear chromosome organization in nematodes, we will be able to conduct future studies to determine the factors responsible for initiating this organization.
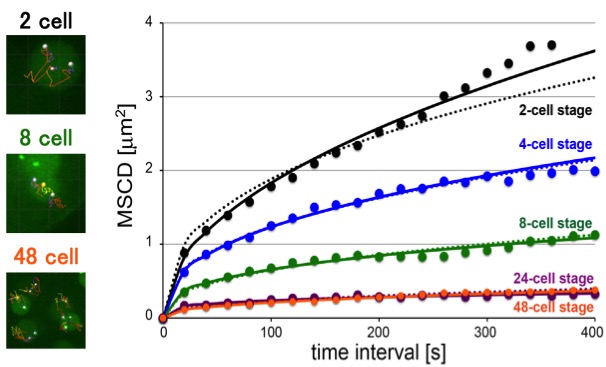
Figure. [Left images] Examples of the tracking (lines) of specific chromosomal loci (white dots) at indicated stages. The yellow dot reveals the center of the nucleus (not shown for the 48-cell stage). [Right graph] MSCD (mean squared change in distance) analyses of mobility: MSCD of the loci was plotted against the time interval (τ) for each stage. Some of the panels are identical as the panels in the paper (Arai et al., 2017)
Enhancer adoption changes limb morphology
![]()
Enhancer adoption caused by genomic insertion elicits interdigital Shh expression and syndactyly in mouse
Kousuke Mouri, Tomoko Sagai, Akiteru Maeno, Takanori Amano, Atsushi Toyoda, Toshihiko Shiroishi
PNAS Published online before print December 18, 2017 DOI:10.1073/pnas.1713339115
Pressrelease (In Japanese only)
Gene expression is regulated by tissue or organ-specific enhancer(s). Acquisition of new enhancers can alter the regulation of developmental genes, and may introduce morphological novelty in evolution. Reconfiguration between a gene and pre-existing enhancers of another gene by chromosomal rearrangement, which is referred to as “enhancer adoption”, is one possible source of new enhancers.
In this study, we re-examined an old mouse mutant named Hammer toe (Hm), which arose spontaneously over a half century ago and exhibits syndactyly with interdigital webbing. We revealed that a 150-kb non-coding genomic fragment that was originally located in chromosome 14 has been inserted into a genomic region upstream to Sonic hedgehog (Shh), located in chromosome 5. ATAC-seq and subsequent reporter assays in vivo revealed that the inserted fragment contains three interdigital enhancers to induce Shh expression in the interdigital regions in Hm. This ectopic Shh upregulates Chordin (Chrd), which in turn inhibits BMP signaling, and eventually results in syndactyly and web formation. Since the donor fragment residing in chromosome 14 has enhancer activity to induce interdigital gene expression, the Hm mutation appears to be an archetypal case of enhancer adoption. Our series of genomic deletion induced by the CRISPR-Cas9 system revealed that three enhancers in 150-kb fragment induce syndactyly in a cooperative manner. Acquisition of the combination of enhancers may have important role for the enhancer adoption.
This study was carried out as a collaboration of Kousuke Mouri, Tomoko Sagai, Akiteru Maeno, Takanori Amano and Toshihiko Shiroishi of Mammalian Genetics Laboratory, and Atsushi Toyoda of Comparative Genomics Laboratory, NIG. This study was supported by JSPS KAKENHI JP15J06985, JP17K15162 and JP17K19411.
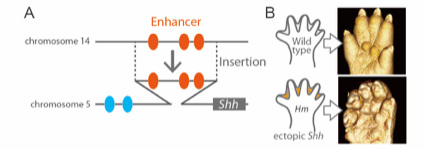
Fig. 1 (A) Hm has an insertion into the upstream region of Shh gene. Inserted fragment includes three interdigital enhancers. (B) X-ray micro-CT images of limb (P0). Shh is ectopically expressed in the interdigital region, and causes interdigital webbing in Hm.
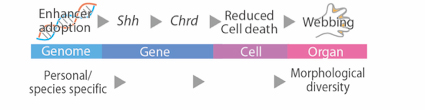
Fig. 2 The process that the morphological alteration in Hm was caused by the enhancer adoption.
You can see the several data of micro CT-scan which is one of the methods for this research below.
- NIG 3d Imaging Room(Facebook)
- 3D Imaging Room in NIG(sketchfab)
You can read the short review for this article.
You can read our news release in EurekAlert! about this article.
Protocadherin-αC2 is required for diffuse projections of serotonergic axons
Division of Neurogenetics / Iwasato Group
Protocadherin-αC2 is required for diffuse projections of serotonergic axons
Shota Katori, Yukiko Noguchi-Katori, Atsushi Okayama, Yoshimi Kawamura, Wenshu Luo, Kenji Sakimura, Takahiro Hirabayashi, Takuji Iwasato & Takeshi Yagi
Scientific Reports, 7, Article number: 15908 (2017) DOI:10.1038/s41598-017-16120-y
Serotonergic axons extend diffuse projections throughout various brain areas, and serotonergic system disruption causes neuropsychiatric diseases. Loss of the cytoplasmic region of protocadherin-α (Pcdh-α) family proteins, products of the diverse clustered Pcdh genes, causes unbalanced distributions (densification and sparsification) of serotonergic axons in various target regions. However, which Pcdh-α member(s) are responsible for the phenotype is unknown. Here we demonstrated that Pcdh-αC2 (αC2), a Pcdh-α isoform, was highly expressed in serotonergic neurons, and was required for normal diffusion in single-axon-level analyses of serotonergic axons. The loss of αC2 from serotonergic neurons, but not from their target brain regions, led to unbalanced distributions of serotonergic axons. Our results suggest that αC2 expressed in serotonergic neurons is required for serotonergic axon diffusion in various brain areas. The αC2 extracellular domain displays homophilic binding activity, suggesting that its homophilic interaction between serotonergic axons regulates axonal density via αC2’s cytoplasmic domain.
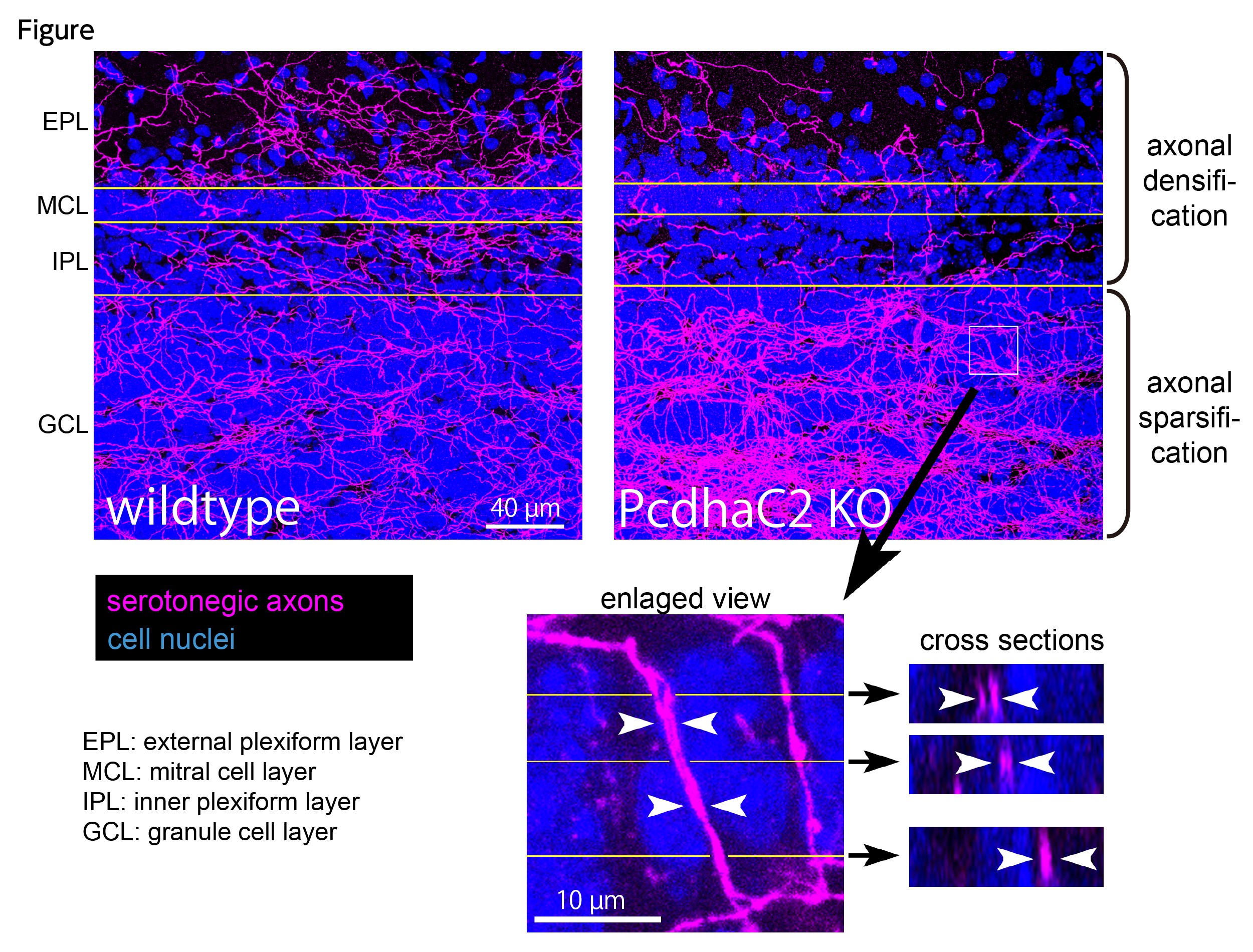
Figure. Serotonergic axons in the olfactory bulb. Wildtype mice (left) show almost uniform distributions in serotonergic axons (magenta) in all layers. In contrast, protocadherin-α C2 deficient mice (right) exhibit unbalanced serotonergic-axon distributions (densification in granule cell layer and sparsification in the other layers). In the granule cell layer (box), fasciculating serotonergic axons are observed.
Source: Scientific Reports 7 Article number: 15908(2017)
DOI: 10.1038/s41598-017-16120-y
Revisiting the ‘Chemotropic Theory’ of neural wiring
Division of Brain Function / Hirata Group
Netrin-1 Derived from the Ventricular Zone, but not the Floor Plate, Directs Hindbrain Commissural Axons to the Ventral Midline
Kenta Yamauchi, Maya Yamazaki, Manabu Abe, Kenji Sakimura, Heiko Lickert, Takahiko Kawasaki, Fujio Murakami, and Tatsumi Hirata
Scientific Reports, DOI:10.1038/s41598-017-12269-8
In this study, we have provided evidence that Netrin-1 (Ntn1) from the ventricular zone (VZ), but not the floor plate (FP), a glial structure occupying the ventral midline (VM), guides hindbrain commissural axon (CA) to the VM. Our results are incompatible with the prevailing view that Ntn1 is a chemoattractant for CAs, rather suggest a novel mechanism that VZ-derived Ntn1 directs CAs to the VM by its local actions.
More than a century ago, based on his observations of CA growth towards the VM, Ramón y Cajal proposed the ‘chemotropic theory’, attraction of growth cones by a long-range diffusible cue emanating from their targets. Ntn1 emanating from the VM has been assumed as the chemoattractant. Consistent with this view, Ntn1 is expressed in the VM and can attract CAs at a distance in vitro. However, Ntn1 is expressed in the vicinity of the CA path, the VZ, in addition to the FP, raising an alternative possibility that Ntn1 of extra-FP origin directs CAs to the VM. To test this possibility, we generated Ntn1 FP conditional mutant (Ntn1FP-Ko) and VZ conditional mutant mice (Ntn1VZ-Ko). If Ntn1 acts CAs as a long-range diffusible chemoattractant, CA guidance to the VM should be disrupted in Ntn1FP-Ko mice. Contrary to the prediction, deletion of Ntn1 from the VZ highly disrupted CA guidance to the VM, whereas deletion of Ntn1 from the FP had little impact on it (Figure). Our results fail to support the chemotropic theory proposed by Ramón y Cajal, and rather suggest that local actions of Ntn1 from the VZ direct CAs to the VM.
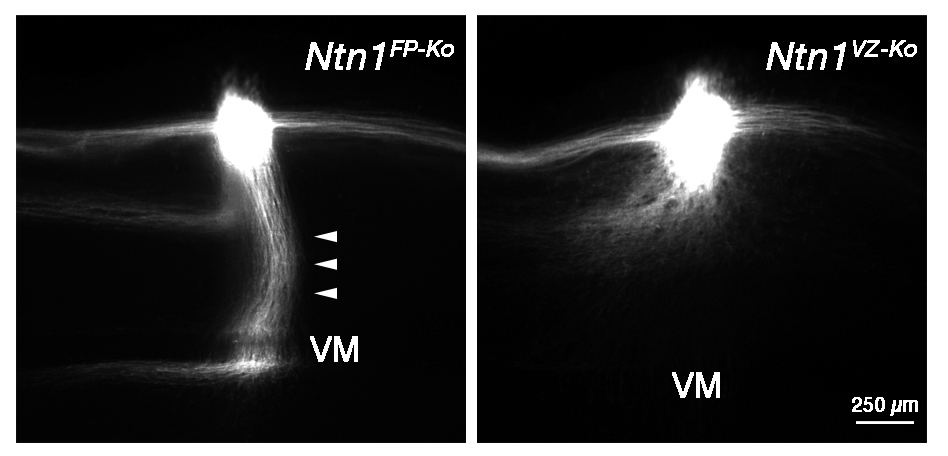
Hindbrain CA growth in Ntn1FP-Ko and Ntn1VZ-Ko mice. Deletion of Ntn1 from the VZ highly disrupts CA guidance to the VM (right), whereas deletion of Ntn1 from the FP has little impact on it (left, arrowheads). CA is labeled with a fluorescent lipophlic dye, DiI.
A new laboratory established in the Center for Frontier Research
Fumi KUBO joined the Center for Frontier Science as of December 1, 2017.
KUBO, Fumi:Center for Frontier Research,Systems Neuroscience Laboratory

- KUBO, Fumi
Associate Professor
Center for Frontier Research is an incubation center to simultaneously develop two elements: human resources and new research fields. Promising young scientists conduct research as principal investigator (tenure-track associate professor) to explore new frontiers in genetics and related areas, taking advantage of NIG’s research infrastructure and various support systems.















