Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
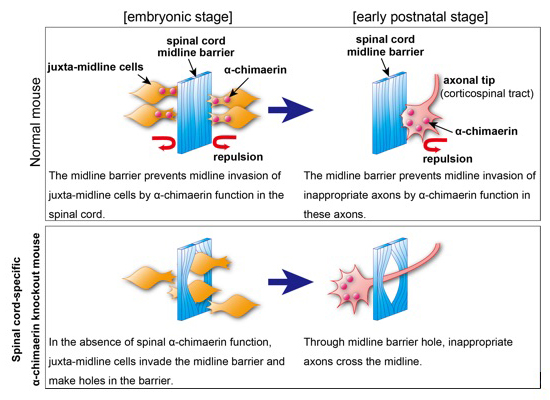
Spinal RacGAP α-chimaerin is required to establish the midline barrier for proper corticospinal axon guidance
Press release
Spinal RacGAP α-chimaerin is required to establish the midline barrier for proper corticospinal axon guidance
Shota Katori, Yukiko Noguchi-Katori, Shigeyoshi Itohara, Takuji Iwasato
Journal of Neuroscience 26 July 2017, 3123-16; DOI:10.1523/JNEUROSCI.3123-16.2017
Pressrelease (In Japanese only)
The midline barrier plays a critical role in midline axon guidance, which is fundamental to the formation of neural circuits that are responsible for proper left-right coordination of our body. Studies have revealed some of the mechanisms underlying how the midline barrier navigates axons. In contrast, the establishment of the midline barrier during embryonic development remains unclear. In this study, Katori et al. determined that α-chimaerin is required for the formation of an intact midline barrier. Spinal cord-specific α-chimaerin knockout mice had spinal midline barriers with numerous breaks (holes), through which corticospinal axons aberrantly crossed the midline. Katori et al. propose that α-chimaerin protects the midline barrier by mediating cell repulsive signaling in juxta-midline cells, which prevents these cells from invading the midline.
Source: Journal of Neuroscience 26 July 2017, 3123-16;
DOI:10.1523/JNEUROSCI.3123-16.2017
Systematic identification of regulatory elements derived from human endogenous retroviruses
Division of Human Genetics / Inoue Group
Systematic Identification and Characterization of Regulatory Elements Derived from Human Endogenous Retroviruses.
Jumpei Ito, Ryota Sugimoto, Hirofumi Nakaoka, Shiro Yamada, Tetsuaki Kimura, Takahide Hayano, and Ituro Inoue.
PLoS Genetics. Jul 12;13(7):e1006883. 2017. DOI:10.1371/journal.pgen.1006883
Human endogenous retroviruses (HERVs) have regulatory elements that possibly influence the transcription of host genes. We systematically identified these HERV regulatory elements (HERV-REs) based on publicly available datasets of ChIP-Seq. Overall, 794,972 HERV-REs were identified. Clustering analysis showed that HERVs can be grouped according to the TF binding patterns; HERV groups bounded by pluripotent TFs (e.g., SOX2, POU5F1, and NANOG), endoderm TFs (e.g., GATA4/6, SOX17, and FOXA1/2), hematopoietic TFs (e.g., SPI1, GATA1/2, and TAL1), and CTCF were identified. Regulatory elements of HERVs tended to locate nearby genes involved in immune responses, indicating that the regulatory elements play an important role in controlling the immune regulatory network. Finally, we constructed dbHERV-REs, an interactive database of HERV regulatory elements (http://herv-tfbs.com/). This study provides fundamental information in understanding the impact of HERVs on host transcription, and offers insights into the transcriptional modulation systems of HERVs.
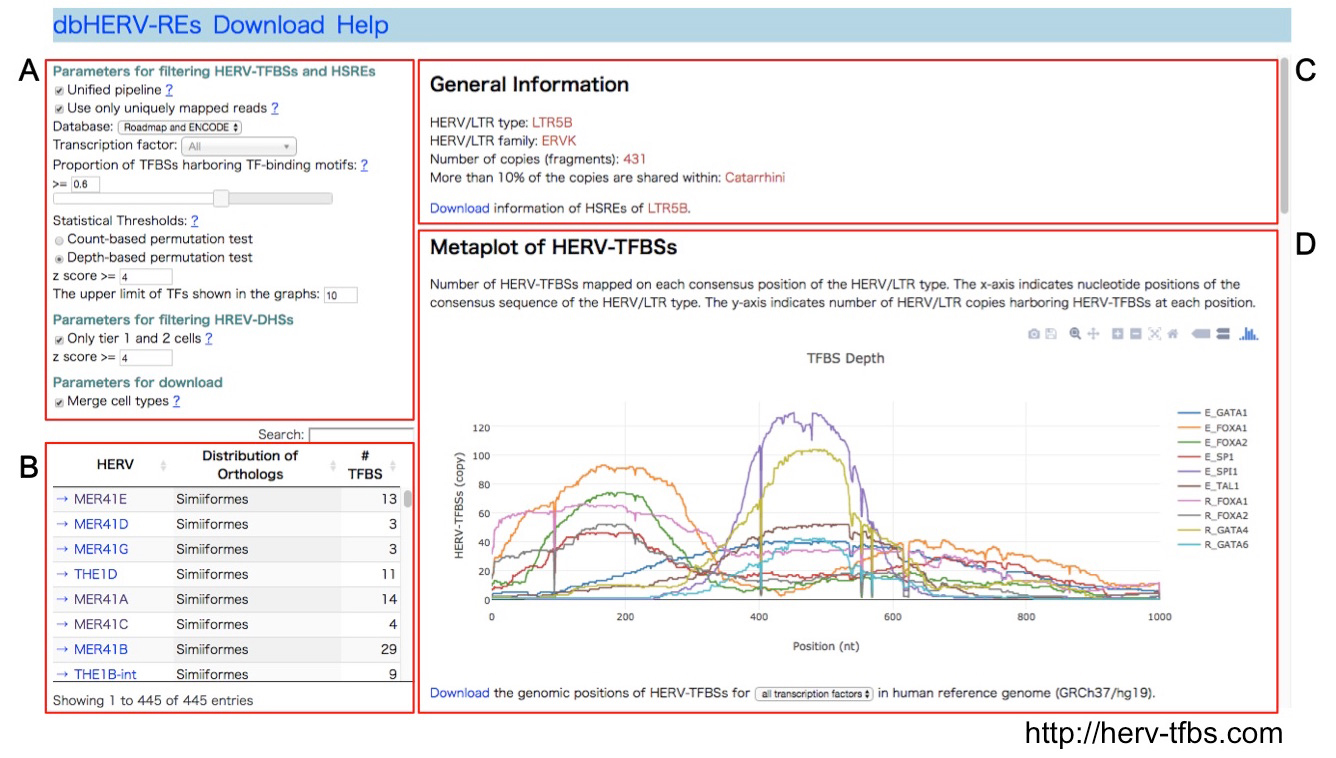
dbHERV-RE (http://herv-tfbs.com/). First, users choose a transcription factor and other parameters (A). Second, users select a type of HERVs (B). dbHERV-REs displays general information of the HERVs (phylogenetic classification, copy number, and insertion date) (C) and visualizes genetic positions of HERV-REs on the HERV sequence and the human reference genome (D).
Dynamic organization of chromatin domains in living cells.
Press release
Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging
Tadasu Nozaki, Ryosuke Imai, Mai Tanbo, Ryosuke Nagashima, Sachiko Tamura, Tomomi Tani, Yasumasa Joti, Masaru Tomita, Kayo Hibino, Masato T. Kanemaki, Kerstin S. Wendt, Yasushi Okada, Takeharu Nagai, and Kazuhiro Maeshima
Molecular Cell Published: July 13, 2017 DOI:10.1016/j.molcel.2017.06.018
Press release (In Japanese only)
The eukaryotic genome is organized within cells as chromatin. For proper information output, higher-order chromatin structures can be regulated dynamically. How such structures form and behave in various cellular processes remains unclear. Here, by combining super-resolution imaging (photoactivated localization microscopy [PALM]) and single-nucleosome tracking, we developed a nuclear imaging system to visualize the higher-order structures along with their dynamics in live mammalian cells. We demonstrated that nucleosomes form compact domains with a peak diameter of ∼160 nm and move coherently in live cells. The heterochromatin-rich regions showed more domains and less movement. With cell differentiation, the domains became more apparent, with reduced dynamics. Furthermore, various perturbation experiments indicated that they are organized by a combination of factors, including cohesin and nucleosome-nucleosome interactions. Notably, we observed the domains during mitosis, suggesting that they act as building blocks of chromosomes and may serve as information units throughout the cell cycle.
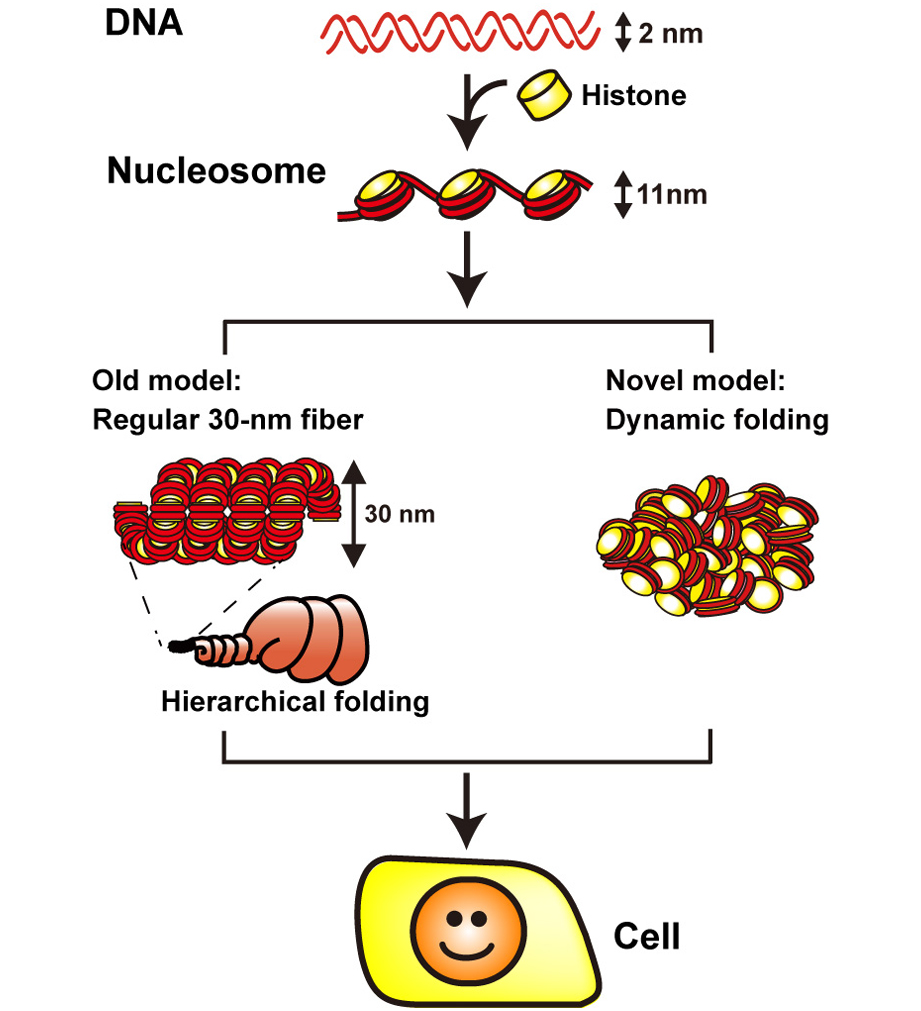
1. DNA (in red) is wrapped around core histone (in yellow) and forms a nucleosome structure (or 10-nm fiber). In the old model (left), the nucleosome fiber had long been assumed to fold into a 30-nm chromatin fiber, and subsequently into helically folded larger fibers (Hierarchical folding). In new current model (right), chromatin is composed of irregularly folded 10-nm fibers, without 30-nm chromatin fibers (dynamic folding) and stored in the cell.
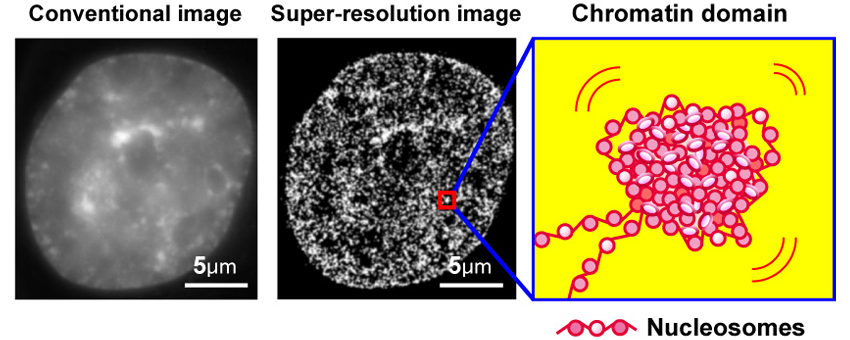
2. Left, conventional DNA staining image; center, super-resolution image of chromatin; a model of chromatin domain.
What makes domestic animals such as mice and dogs tame
Press release
Selective breeding and selection mapping using a novel wild-derived heterogeneous stock of mice revealed two closely-linked loci for tameness
Yuki Matsumoto, Tatsuhiko Goto, Jo Nishino, Hirofumi Nakaoka, Akira Tanave, Toshiyuki Takano-Shimizu, Richard F Mott, Tsuyoshi Koide
Scientific Reports 7, Article number: 4607 (2017) DOI:10.1038/s41598-017-04869-1
Pressrelease (In Japanese only)
Tameness play important role during the process of domestication, and can be divided into two potential components: motivation to approach humans (active tameness) and reluctance to avoid them (passive tameness). To understand the genetic basis associated with active tameness in mice we applied selective breeding of a genetically heterogeneous population that we founded by crossing eight wild mouse strains. As a result of selective breeding, the level of active tameness increased over the generations, compared to an unselected control experiment. We performed two selection and two control experiments. Genetic differences between the selected and control groups, measured using a high-dense array of single-nucleotide polymorphisms, were assessed using a computer simulation experiment. In one selection experiment we found a significant increase in the occurrence of a particular genomic segment present in just one of the founder strains, compared to the control groups. This selected region contains two loci related to active tameness and is syntenic to genomic regions which are known to be a region selected during dog domestication, suggesting that responsible genes in these loci are associated with active tameness in both mouse and dog.
A new laboratory established in the Center for Frontier Research
Yasuto MURAYAMA joined the Center for Frontier Science as of July 1, 2017.
MURAYAMA, Yasuto:Center for Frontier Research,Chromosome Biochemistry Laboratory

- MURAYAMA, Yasuto
Associate Professor
Center for Frontier Research is an incubation center to simultaneously develop two elements: human resources and new research fields. Promising young scientists conduct research as principal investigator (tenure-track associate professor) to explore new frontiers in genetics and related areas, taking advantage of NIG’s research infrastructure and various support systems.















