Sensor Residues in Bacterial Condensin That Discriminate Between ssDNA
Niki Group • Microbial Physiology Laboratory
Involvement of the inner surface residues of bacterial SMC protein MukB in the ssDNA binding in vitro
Koichiro Akiyama (秋山光市郎), Koichi Yano (矢野晃一) & Hironori Niki (仁木宏典)
Communications Biology (2025)
Inside cells, DNA that is much longer than the cell itself is compactly stored; for example, Escherichia coli contains approximately 1.5 cm of DNA within a 2-micrometer cell. The condensation of such long DNA is facilitated by ring-shaped proteins called condensins, which play essential roles in both prokaryotic and eukaryotic organisms. In E. coli, the condensin known as MukB mediates chromosome condensation. MukB forms a ring by joining two rod-shaped molecules at both ends and captures DNA within this ring to promote compaction. Notably, MukB captures single-stranded DNA (ssDNA) more efficiently than double-stranded DNA (dsDNA).
This study aimed to elucidate the molecular mechanism underlying this specificity. Multiple MukB variants with single amino acid substitutions were generated and analyzed. Mutations in residues located inside the ring impaired ssDNA binding and reduced capture efficiency. These residues are aligned linearly along the inner surface of the ring, and upon ssDNA capture, they tether the two ends of MukB like a sash, thereby stabilizing the ring structure. This structural stabilization likely explains MukB’s higher affinity for ssDNA.
Since ssDNA is formed in specific genomic regions, the preferential binding of MukB to ssDNA may contribute to its accumulation in these regions. Further studies are needed to clarify the physiological significance of this specificity.
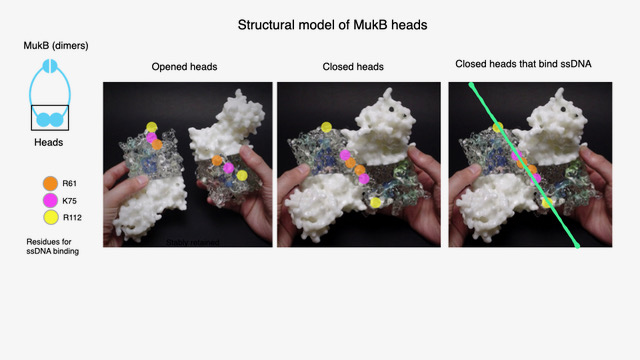
In the closed conformation, the sensor residues align along a single line. When these residues detect a single-stranded DNA molecule inside the ring, the DNA is stably retained.















