Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Does Wnt function like odorants to orient cells?
Sawa Group • Multicellular Organization Laboratory
Differential functions of multiple Wnts and receptors in cell polarity regulation in C. elegans
Hitoshi Sawa , Masayo Asakawa , Takefumi Negishi
Genetics(2026)DOI:10.1093/genetics/iyag004 DOI:10.1101/2025.11.25.690575
Metazoan species possess multiple Wnt ligands and receptor genes that regulate diverse developmental processes. Because these genes often act redundantly, analysis of single-gene mutants does not necessarily reveal the full roles of Wnt signaling. In C. elegans, three Wnt genes (cwn-1, egl-20, and cwn-2) and three receptor genes (lin-17/Fzd, mom-5/Fzd, and cam-1/Ror) redundantly regulate the polarity of asymmetrically dividing seam cells. Here, we comprehensively analyzed genetic interactions among these Wnt and receptor genes. In mom-5 mutant backgrounds, additional mutations in Wnt genes disrupted cell polarization. In contrast, in cam-1 mutant backgrounds, Wnt mutations frequently caused abnormal polarity orientation. These findings indicate that MOM-5 and CAM-1 play distinct roles in establishing cell polarization and determining its orientation, respectively. lin-17 mutations suppressed polarity reversal in multiple Wnt compound mutants, suggesting that LIN-17 may function as a molecular switch for polarity orientation. Although all three Wnt genes regulate polarity orientation in a gradient-independent manner in the absence of receptor mutations, in lin-17 mutant backgrounds, reversing the expression gradients of cwn-1 and egl-20, but not cwn-2, enhanced polarity reversal. This suggests that cwn-1 and egl-20 act not only permissively but also instructively to regulate polarity orientation. Together, our results reveal distinct and cooperative functions of multiple Wnt ligands and receptors that ensure robust control of cell polarity.
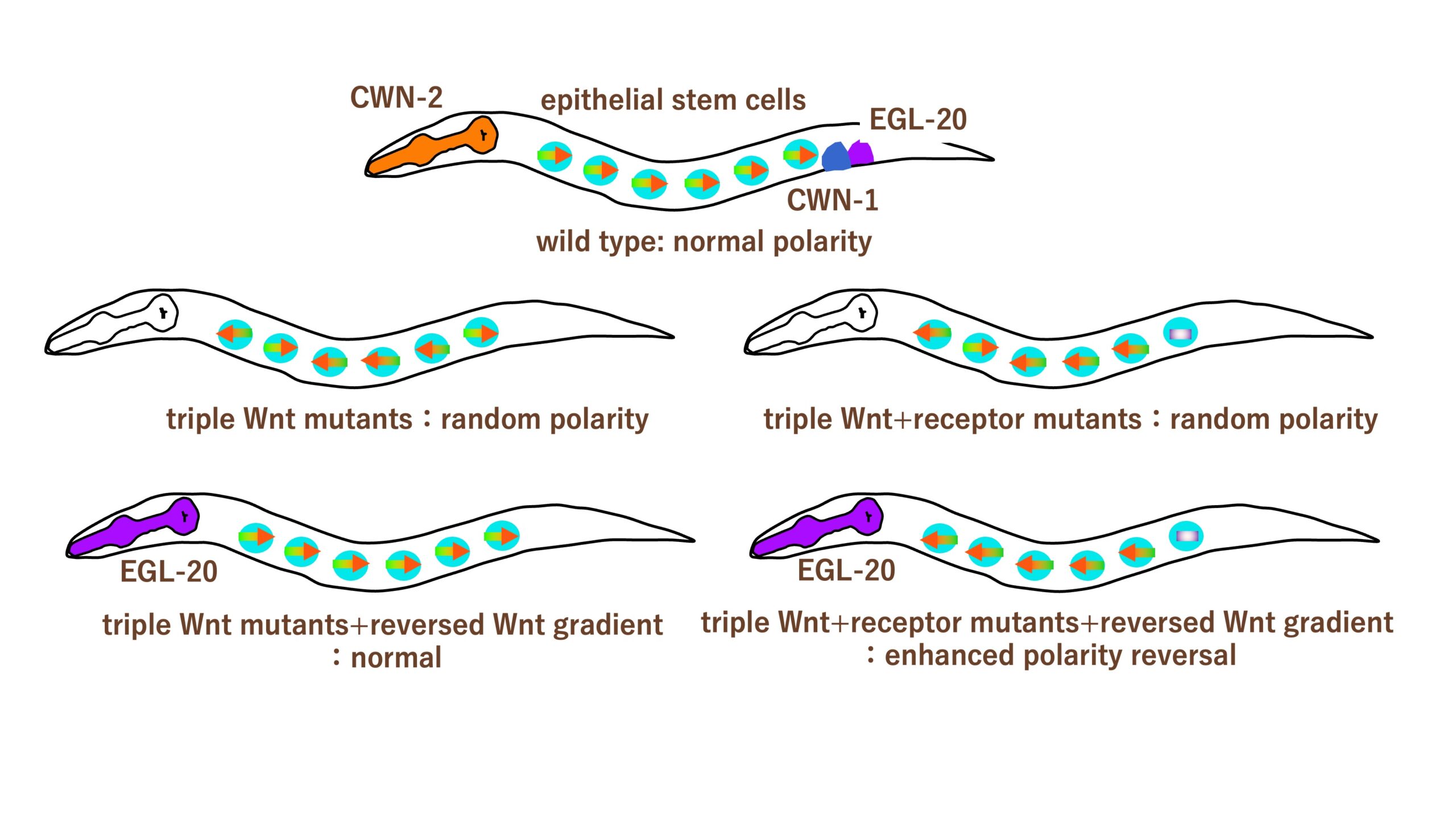
In triple Wnt mutants or triple Wnt+ receptor (lin-17/Frizzled) mutants, polarity of epithelial stem cells are randomized. When Wnt (EGL-20) which is normally expressed posteriorly, is expressed anteriorly reversing the Wnt gradient, polarity defects are rescued in triple Wnt mutants, while it enhances polarity reversal in triple Wnt+ receptor mutants.
Evolving fish in an urban lake──Rapid evolution of stickleback in an urban lake: natural selection driving evolution in real time
Kitano Group • Ecological Genetics Laboratory
Yamasaki, Y.Y., Yamaguchi, R., Nagano, A. J., Chen, B.J., Musto, N., Archambeault, S., Peichel, C.L., Schulien, J.A., Code, T.J., Beauchamp, D.A., and Kitano, J.
Inferring the strength of directional selection on armor plates in Lake Washington stickleback while accounting for migration and drift.
Evolution in press (2025) DOI:10.1093/evolut/qpaf254
Is evolution too slow to observe? Evolution is often thought of as a slow process unfolding over vast geological timescales. Researchers have shown that stickleback fish in Lake Washington (Seattle, USA) have undergone measurable evolutionary change between 2005 and 2022.
The study revealed that sticklebacks with complete bony plates had survival rates several percentage points higher than those with reduced plates, indicating ongoing natural selection. Moreover, the strength of selection appears to have intensified between 2016 and 2022. These findings demonstrate that natural selection can drive rapid evolution in natural populations. The paper was published in Evolution on 16 December 2025.
To rigorously demonstrate that these changes reflect natural selection rather than demographic processes, it is essential to quantify selection while accounting for migration from other populations and genetic drift. Although recent advances in molecular genetics have enabled researchers to identify genes underlying phenotypic traits and to estimate the strength of natural selection acting on them, estimating selection in natural populations remains challenging when migration from other populations occurs, as migration can obscure the effects of selection.
A research group led by Professor Jun Kitano and Assistant Professor Yo Yamasaki of the National Institute of Genetics, together with Assistant Professor Ryo Yamaguchi of Hokkaido University, and collaborators in the United States and Switzerland, integrated whole-genome sequencing data to estimate key population demographic parameters, including migration rates and effective population size. By incorporating these parameters into predictive models, the team was able to quantitatively estimate the strength of natural selection in a natural system. Their analyses further showed that selection pressure increased between 2016 and 2022.
Professor Jun Kitano commented:
“About 20 years ago, I found that sticklebacks in Lake Washington had evolved since the 1960s. When I revisited this lake population in 2022, I was surprised to see that evolution was still ongoing and that selection pressure had intensified. Because Lake Washington is connected to the sea and inflowing rivers, migration from surrounding populations makes it difficult to accurately estimate selection. We have now established a quantitative framework to overcome this challenge. By studying cases of rapid evolution in nature, we can directly observe how natural selection drives evolutionary change in real time.”
This research was supported by JSPS KAKENHI (22H04983, 20J01503, 21H02542, 22KK0105), JST CREST (JPMJCR20S2), and the King County WRIA8 Cooperative Watershed Management Grant Program.
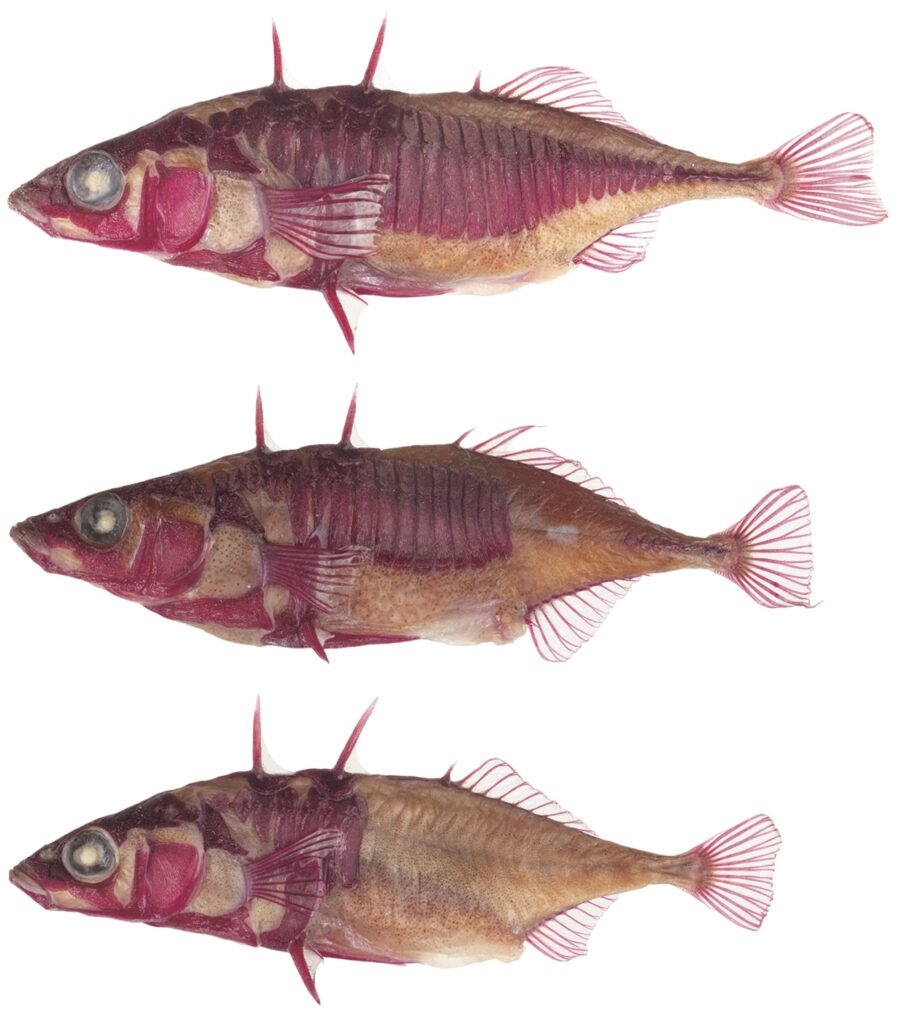
Figure: In Lake Washington, the frequency of stickleback fish with “complete” plates, characterized by bony lateral plates covering the entire flank of the body (top), has increased.
A Method for Precise On–Off Control of Gene Expression in Microalgae
Miyagishima Group • Symbiosis and Cell Evolution Laboratory
IPTG- and estradiol-inducible gene expression systems in the unicellular red alga Cyanidioschyzon merolae.
Takayuki Fujiwara, Shunsuke Hirooka, Shota Yamashita, and Shin-ya Miyagishima.
Plant Physiology(2025) DOI:10.1093/plphys/kiaf575
The genetically tractable unicellular red alga Cyanidioschyzon merolae has a remarkably simple genome (4,775 nucleus-encoded proteins) and cellular architecture. It contains only a single set of most membranous organelles, making it a valuable tool for elucidating the fundamental mechanisms of photosynthetic eukaryotes. However, as in other genetically tractable eukaryotic algae, previously developed systems for inducible gene expression rely on environmental stimuli such as heat shock or ammonium depletion, which impact cellular physiology and thus limit their usage. To overcome this issue, we developed IPTG- and estradiol-inducible gene expression systems in C. merolae in which the addition of these chemicals itself has no impact on cellular growth or the transcriptome. Additionally, we established IPTG- and estradiol-inducible protein knockdown systems and successfully degraded the endogenous chloroplast division protein DRP5B using the estradiol-inducible system. These systems facilitate functional genomic analyses in C. merolae, especially for understanding physiological mechanisms and their interactions in photosynthetic eukaryotes.
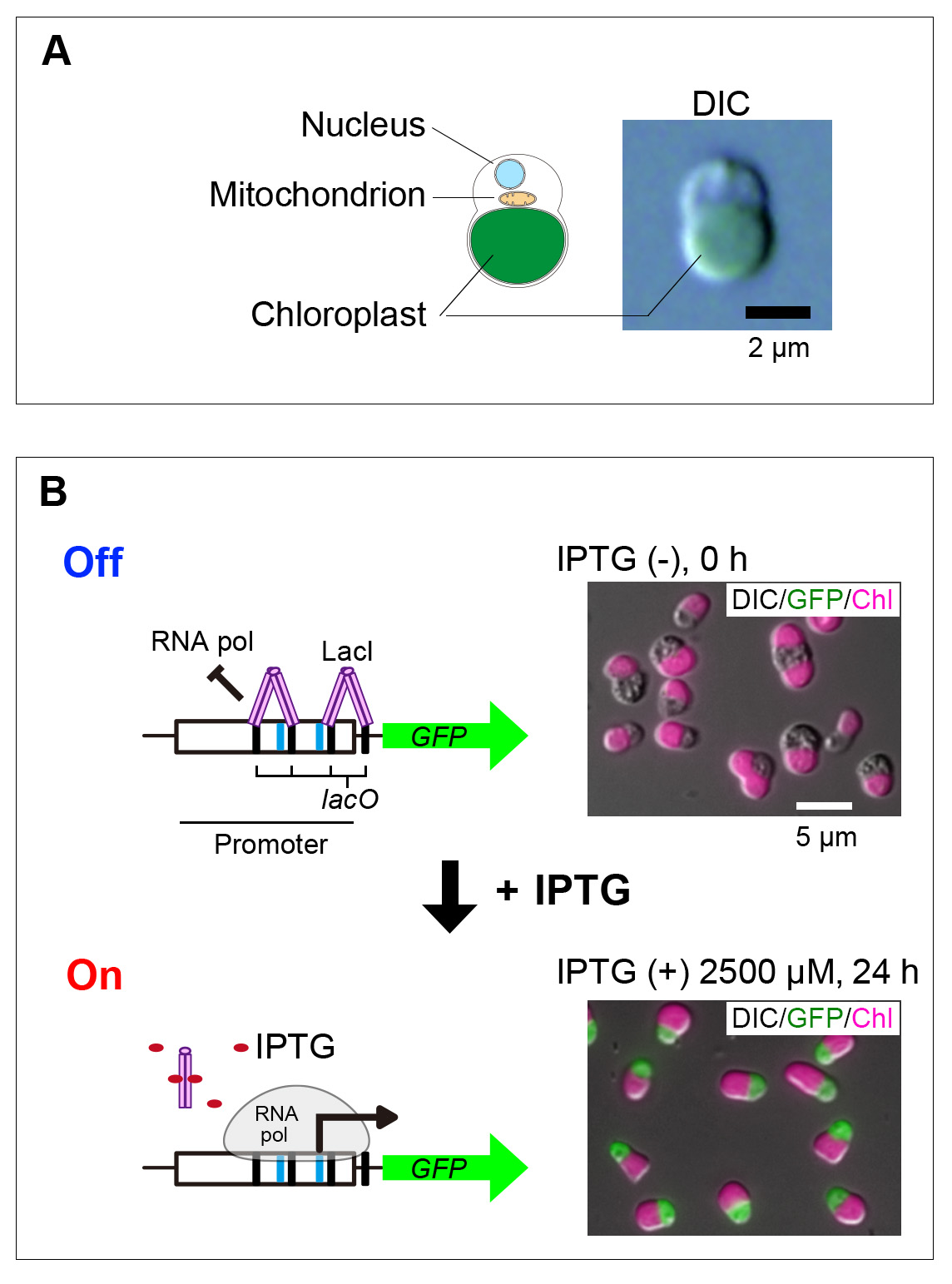
IPTG-inducible gene expression system.
(A) Schematic illustration of the cellular structure and differential interference contrast (DIC) image of Cyanidioschyzon merolae, a species of microalgae.
(B) Schematic overview of the IPTG-inducible gene expression system and fluorescence microscopy images of cells expressing the reporter protein GFP (green) in an IPTG-dependent manner. Four lac operator (lacO) sequences were inserted into the promoter region, and an Escherichia coli–derived LacI repressor was expressed separately to suppress transcription by binding to these sites. Upon addition of IPTG, LacI dissociates from the lacO sequences, allowing transcription to initiate. GFP protein expression was detected from 4 hours after IPTG addition. DIC images show cell outlines, GFP (green) indicates cytosolic fluorescence, and Chl (magenta) represents chloroplast autofluorescence. Scale bars are indicated in the images.
Speciation by sexual selection and hybridization
Kitano Group • Ecological Genetics Laboratory
The interplay of sexual selection and hybridisation can drive sexual radiation
Kotaro Kagawa
Proceedings of the Royal Society B (2025) DOI:/10.1098/rspb.2025.2605
Rapidly diverging lineages often involve interspecific variation in mating displays—such as nuptial coloration, mating songs, and courtship dances. Currently, the evolutionary processes underlying the formation of such groups that exhibit diverse mating displays remain largely unclear, especially when they lack clear ecological differences and significant hybrid incompatibility. Based on computer simulations of evolution, we propose that hybridisation between genetically distinct lineages can provide a route to rapid speciation accompanied by diversification of mating displays. A key process in this mechanism is an alteration of the sexual selection regime, caused by hybridisation increasing genetic variation in mate preference within populations.
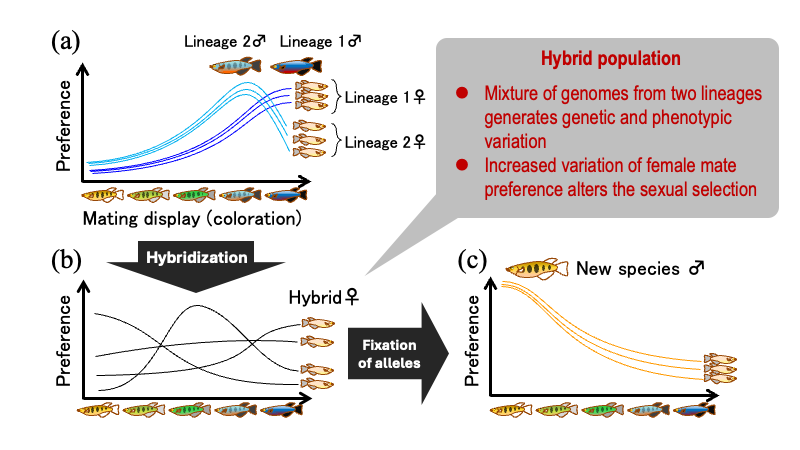
Schematic illustration of our hypothesis. The hypothesis considers hybridization between two genetically distinct lineages within an ancestral species (a). Hybridization generates interindividual variation in female mate preferences, thereby altering the sexual selection regime (b). This mechanism can promote the evolution of a new species with a novel mating display.















