Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Cytoplasmic RNA granule required for regulation of meiosis transition in rice
Nonomura Group / Plant Cytogenetics Laboratory
Impact of protein domains on the MEL2 granule, a cytoplasmic ribonucleoprotein complex maintaining faithful meiosis progression in rice.
Manaki Mimura, Seijiro Ono, Harsha Somashekar, Ken-Ichi Nonomura
New Phytologist 2024 Jul 24. DOI:10.1111/nph.19968
Cytoplasmic ribonucleoprotein (RNP) granules are membraneless structures composed of various RNAs and proteins that play important roles in post-transcriptional regulation. While RNP granules are known to regulate the meiotic entry in some organisms, little is known about their roles in plants.
In this study, we observed the cytoplasmic granular structures of rice RNA-binding protein MEL2, which contributes to the control of meiotic entry timing, in leaf protoplasts and spore mother cells. Our results indicated that MEL2 granules colocalized with processing body and stress granule factors. The maintenance of granule properties modulated by LOTUS domain and the intrinsically disordered region (IDR) is essential for proper MEL2 function in meiosis progression (Figure). MEL2-like proteins widely found in plant kingdom conserved LOTUS domain followed by the IDR despite their diverse domain structures, suggesting the functional conservation of these domains among plant species.
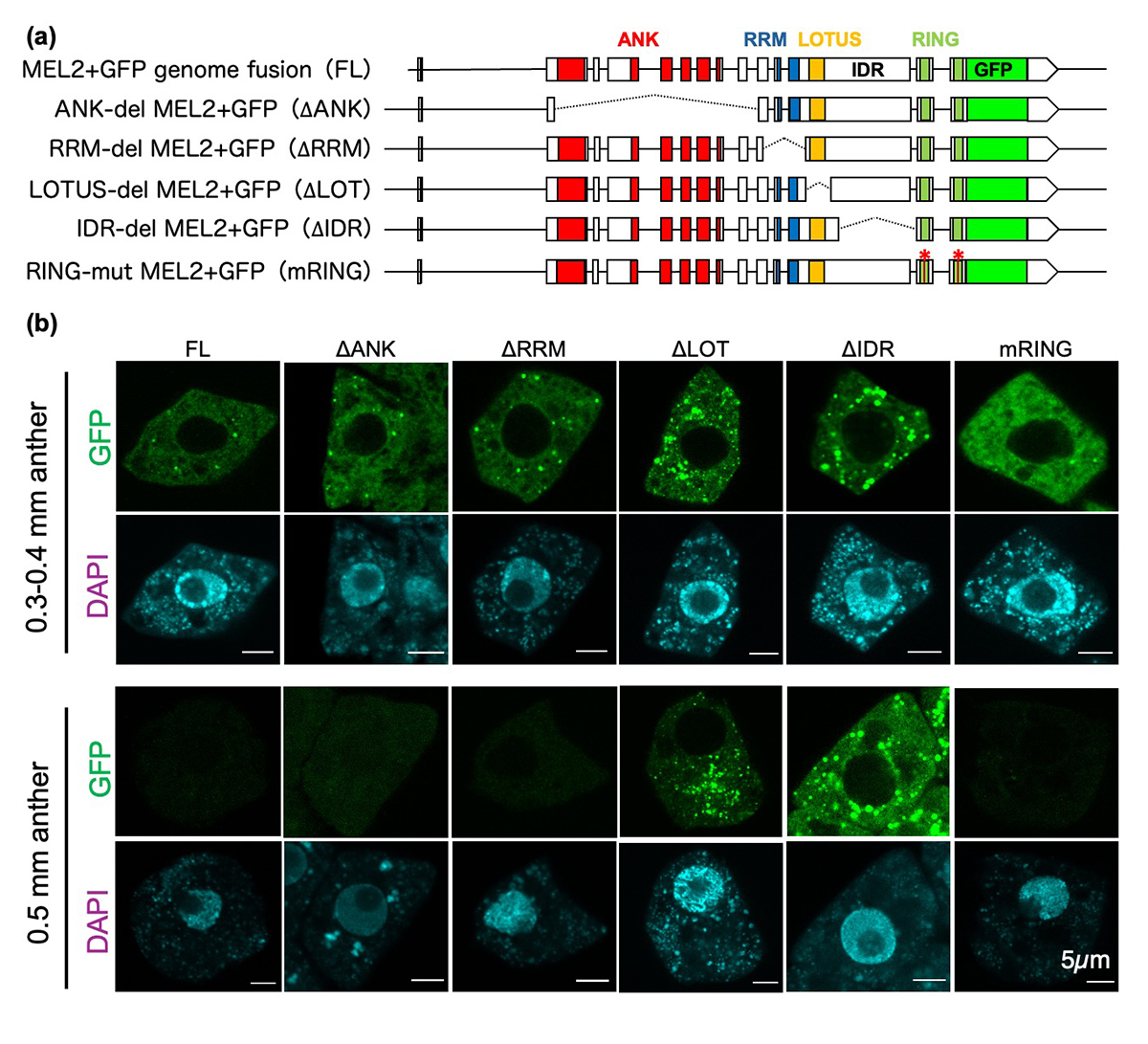
Figure: Roles of MEL2 functional domains in granule formation
(a) Genomic fusion constructs of MEL2 and GFP genes, and its derivatives with deletions (dashed lines) or mutations (asterisks) in MEL2 domains.
(b) MEL2 granule formation in spore mother cells in rice anthers transformed with the constructs in (a). The wildtype MEL2 (FL) formed granules (green) at premeiosis (0.3-0.4mm anthers), and disappeared by meiosis (0.5mm anthers). The mRING failed granule formation, and the granules of ∆LOTUS and ∆IDR were aberrantly carried over to meiocytes, all resulting in defective meiosis and pollen development.
The First Dimerization-Dependent Protein Knockdown Method in Photosynthetic Organisms
Miyagishima Group / Symbiosis and Cell Evolution Laboratory
Development of a rapamycin-inducible protein-knockdown system in the unicellular red alga Cyanidioschyzon merolae.
Takayuki Fujiwara, Shunsuke Hirooka, Shota Yamashita, Fumi Yagisawa and Shin-ya Miyagishima
Plant Physiology (2024) kiae316 DOI:10.1093/plphys/kiae316
An inducible protein-knockdown system is highly effective for investigating the functions of proteins and mechanisms essential for the survival and growth of organisms. However, this technique is not available in photosynthetic eukaryotes. The unicellular red alga Cyanidioschyzon merolae possesses a very simple cellular and genomic architecture and is genetically tractable but lacks RNA interference machinery. In this study, we developed a protein-knockdown system in this alga. The constitutive system utilizes the destabilizing activity of the FK506-binding protein 12 (FKBP12)-rapamycin-binding (FRB) domain of human target of rapamycin kinase or its derivatives to knock down target proteins. In the inducible system, rapamycin treatment induces the heterodimerization of the human FRB domain fused to the target proteins with the human FKBP fused to S-phase kinase associated protein 1 or Cullin 1, subunits of the SCF E3 ubiquitin ligase. This results in the rapid degradation of the target proteins through the ubiquitin-proteasome pathway. With this system, we successfully degraded endogenous essential proteins such as the chloroplast division protein dynamin-related protein 5B and E2 transcription factor, a regulator of the G1/S transition, within 2 to 3 h after rapamycin administration, enabling the assessment of resulting phenotypes. This rapamycin-inducible protein-knockdown system contributes to the functional analysis of genes whose disruption leads to lethality.
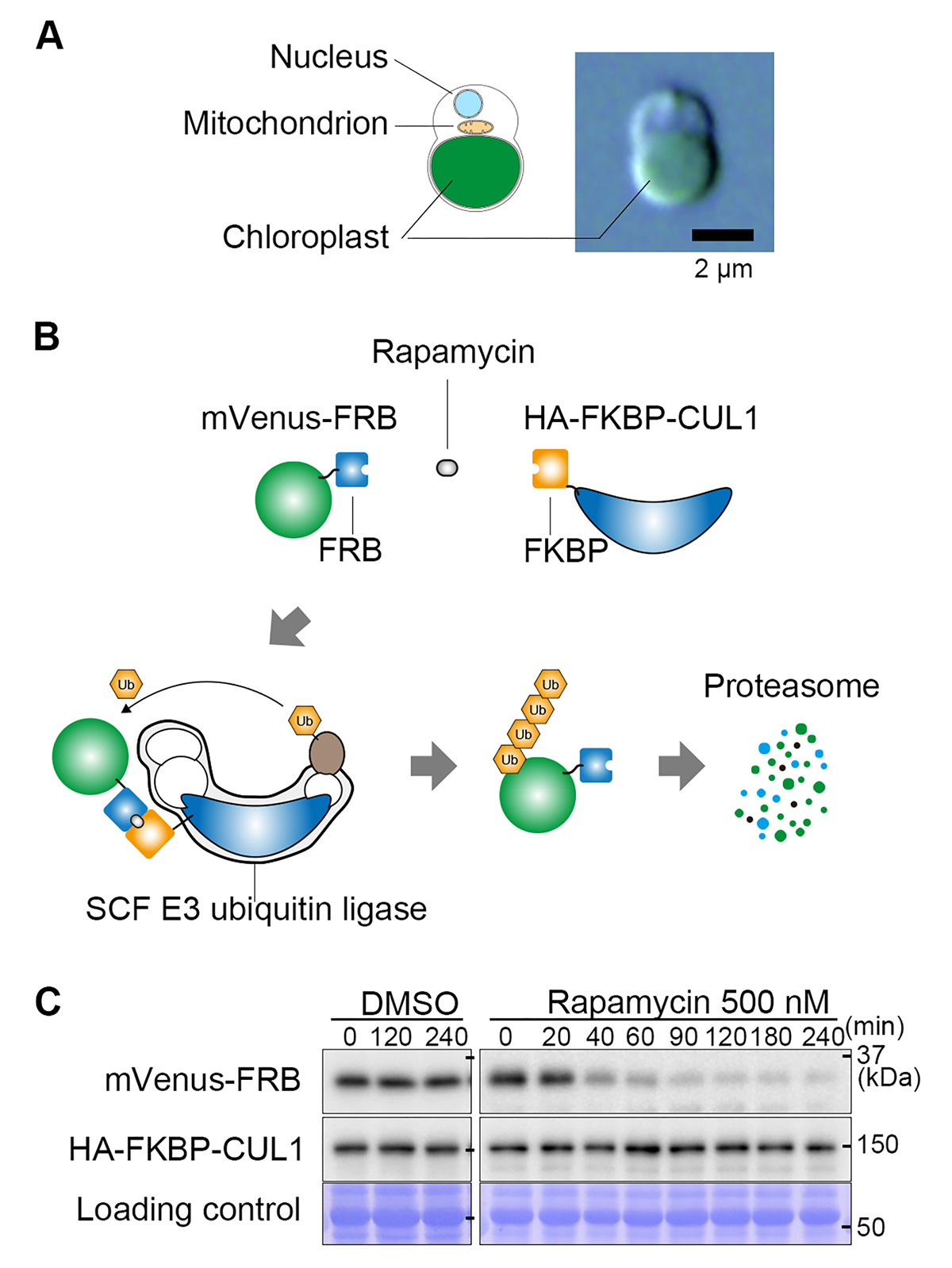
Figure: A rapamycin-inducible protein knockdown system.
(A) The cellular structure of the unicellular red alga Cyanidioschyzon merolae and a differential interference contrast microscopy image.
(B) Mechanism of the rapamycin-inducible protein knockdown system. FRB tags and FKBP proteins form a heterodimer in the presence of rapamycin. The target protein fused with an FRB tag (in this figure, the fluorescent protein mVenus) and the CUL1 subunit of ubiquitin ligase fused with FKBP bind in the presence of rapamycin, resulting in the ubiquitination of FRB-mVenus. The ubiquitinated FRB-mVenus is rapidly degraded by the proteasome.
(C) Immunoblot analysis showing the decrease in mVenus-FRB protein level after the addition of rapamycin.
Summer Holiday (Aug. 15-16)
NIG will be closed from August 15 to August 16, 2024 for summer holiday.
Thank you for your understanding and cooperation.
NIG website service will be stopped several times. July 11 Thu, 00:00 – July 11 Thu, 08:00.
Due to the facility maintenance, the NIG website service will be stopped several times on the following time.
Thank you for your understanding and cooperation.
Time and Date: July 11 Thu, 00:00 – July 11 Thu, 08:00.
以下の期日において、遺伝研公式ウェブサイトの断続的な停止を伴う保守作業が予定されております。
皆様のご理解とご協力をお願いいたします。
停止日時: 7月11日(木) 00:00~7月11日(木) 08:00
理由: 保守作業のため
Problems in Sending and Receiving NIG E-mails between 12:45-13:45 on July 4
Due to the test of the NIG mail system, NIG account mails may not be able to send or receive between 12:45-13:45 on July 4.
We apologize for any inconvenience and appreciate your understanding and cooperation.
Where are the replication initiation zones located in mammalian genome? (Review paper)
Kanemaki Group / Molecular Cell Engineering Laboratory
Replication initiation sites and zones in the mammalian genome: Where are they located and how are they defined?
Xiaoxuan Zhu, Masato T. Kanemaki*
*Corresponding author
DNA Repiar (2024) 141, 103713 DOI:10.1016/j.dnarep.2024.103713
The question of how genomic DNA is replicated is one of the key themes in molecular biology, as abnormalities in DNA replication are related to genome instability, genetic disorders, cell death, and cancer. Therefore, DNA replication has been extensively studied over the years using Escherichia coli and budding yeast as model organisms. In these unicellular models, DNA replication starts from genomic regions with consensus DNA sequences. On the other hand, in mammalian cells, including humans, consensus sequences appear to be absent, and it is still not well understood where DNA replication begins and how are they defined in the genome. Hence, this review paper compiles and compares the results of large-scale high-throughput sequencing-based explorations for mapping initiation sites/zones reported so far. Furthermore, it highlights the similarities and differences in the DNA replication mechanisms between budding yeast and mammalian cells, presents a hypothetical mechanism for how replication initiation zones are defined in mammalian cells, and discusses the future directions and challenges in DNA replication research.
The first author, Dr Xiaoxuan Zhu, has been supported as an NIG post-doctoral fellow.
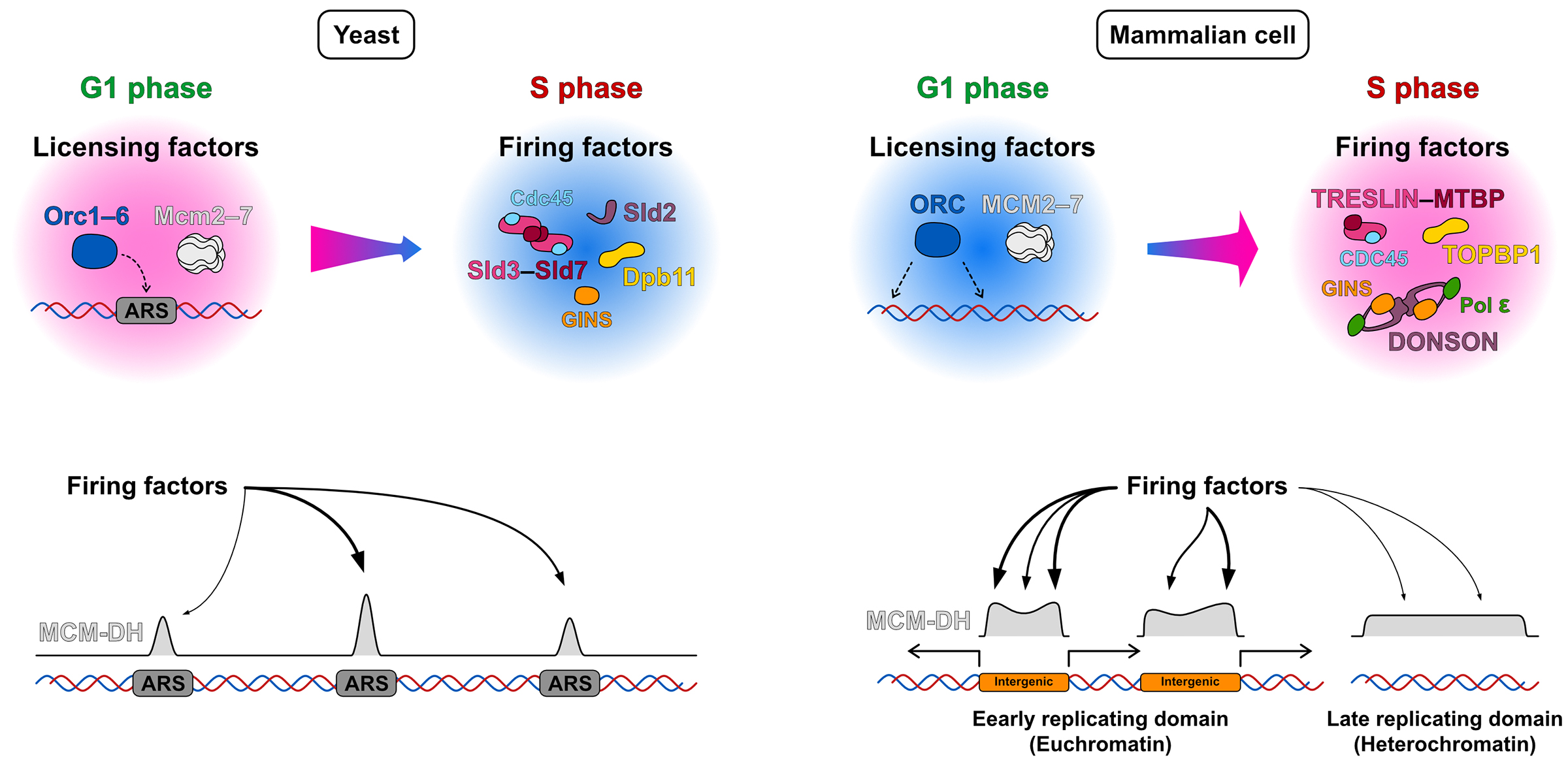
Figure: Differences in the mechanisms of replication initiation sites/zones selection between budding yeast and mammalian cells: In yeast, licensing factors accumulate at ARS (autonomously replicating sequence) sites containing a consensus sequence, leading to replication initiation at these ARS sites. In contrast, in mammalian cells, licensing factors are widely distributed across the genome. It is thought that the subsequent accumulation of firing factors at specific locations is crucial for forming initiation zones.















