Behaviors of nucleosomes with mutant histone H4s
Maeshima Group / Genome Dynamics Laboratory
Kurokawa Group / Genome Evolution Laboratory
Comparative Genomics Laboratory
Behaviors of nucleosomes with mutant histone H4s in euchromatic domains of living human cells.
Adilgazy Semeigazin, Shiori Iida, Katsuhiko Minami, Sachiko Tamura, Satoru Ide, Koichi Higashi, Atsushi Toyoda A, Ken Kurokawa, Kazuhiro Maeshima *
* corresponding author
Histochemistry and Cell Biology 2024 May 14 DOI:10.1007/s00418-024-02293
DNA is a negatively charged polymer that wraps around an octamer of basic core histone proteins through electrostatic interactions, forming a nucleosome. The core histone octamer consists of eight proteins in total: two each of histones H2A, H2B, H3 and H4. Each histone has a “tail” at its terminus that lacks a specific structure and is subject to various modifications. Histone H2A has two tails, so a single nucleosome has a total of ten histone tails. Acetylation of numerous lysine residues on histone tails neutralizes their positive charges, thereby weakening their attraction to DNA and neighboring nucleosomes and causing loss of nucleosome-nucleosome interactions. A specific acetylation of histone [ histone H4 lysine-16 (H4K16Ac)] has been shown to disrupt the chromatin structure in vitro. However, it was unclear whether specific acetylation of histone tails can change the behavior of nucleosomes with that acetylation in living human cells.
A research team led by Professor Kazuhiro Maeshima of Genome Dynamics Laboratory (NIG), including a SOKENDAI graduate student Adilgazy Semeigazin (MEXT Scholar), SOKENDAI graduate students (former SOKENDAI Special Researchers, JSPS Research Fellows DC2) Shiori Iida and Katsuhiko Minami, Technical Staff Sachiko Tamura, former Assistant Professor Satoru Ide (currently Researcher at Tokyo Metropolitan Institute of Medical Science), together with Assistant Professor Koichi Higashi of Genome Evolution Laboratory (NIG), Professor Atsushi Toyoda of Comparative Genomics Laboratory (NIG), and Professor Ken Kurokawa of Genome Evolution Laboratory (NIG), analyzed the local movement of nucleosomes containing the mutation in the histone H4 tail to mimic deacetylation or acetylation. The results showed that their movement was not affected by the mutations because nucleosomes containing these mutated histones were embedded in condensed chromatin domains.
In this work, the research team ectopically expressed wild-type (wt) or mutated H4s (H4K16 point, H4K5,8,12,16 quadruple, and H4 tail deletion) fused with HaloTag in HeLa cells. Expressed wtH4-Halo, H4K16-Halo mutants, and multiple H4-Halo mutants had the euchromatin-concentrated distribution. Consistently, the genomic regions of the wtH4-Halo nucleosomes corresponded to Hi-C contact domains with active chromatin marks. Utilizing single-nucleosome imaging, the authors found that none of the H4 deacetylation or acetylation mimicked H4 mutants altered the overall local nucleosome motion. Interestingly, H4 with four lysine-to-arginine mutations displayed a substantial freely diffusing fraction in the nucleoplasm, whereas H4 with a truncated N-terminal tail was incorporated in heterochromatic regions as well as euchromatin (Figure). This study indicates the power of single-nucleosome imaging to understand individual histone/nucleosome behavior reflecting differential chemical modification states in living cells.
This year marks the centenary of the discovery of DNA staining by Robert Feulgen. This article was published in a special issue of the US journal Histochemistry and Cell Biology, “The Centennial of the Feulgen Reaction – Imaging the Genome.“
This work was supported by JSPS Fellowship, JSPS grants (24H00061, 21H02453, 20H05936, 22H05606, 23K17398, 21H02535, 23KJ0996, 23KJ0998, 22H04925 (PAGS)), a Japan Science and Technology Agency JST SPRING (JPMJSP2104), and the Takeda Science Foundation.
Video: (Left) Movement of ectopically expressed wild-type (wt) H4-Halo. Most of these were incorporated into nucleosomes and showed fluctuations. (Right) Movement of H4R-quad, a deacetylation-mimetic H4 quad mutant. They were less incorporated into the nucleosomes, with most diffusing freely.
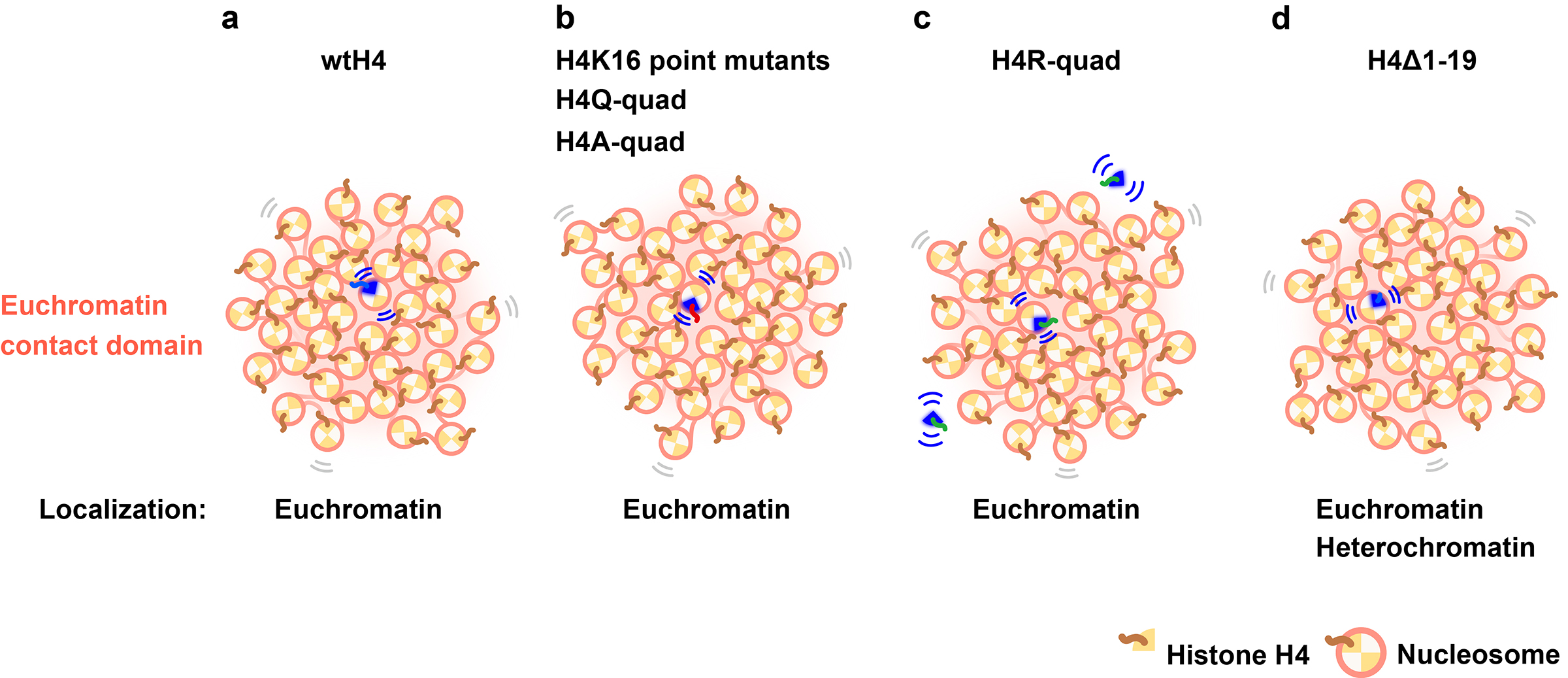
Figure: (a) Ectopically expressed wtH4-Halo was concentrated in the euchromatin contact domain. wtH4-Halo contacts adjacent nucleosomes via an H4 tail. (b) Mutations on the H4 tail (H4K16 point mutants, H4Q-quad, and H4A-quad) did not impact local nucleosome motion because the mutant nucleosomes were embedded in condensed euchromatic domains. Mutations on H4-Halo may have impeded interactions with neighboring nucleosomes, but the surrounding endogenous (wt) H4 nucleosomes were able to maintain contact with the mutant nucleosomes and thus constrain their motion. (c) H4R-quad has a nucleosome incorporation problem and has a large pool of freely diffusing H4R-quad (Movie). However, the incorporated fraction shows a similar motion as wtH4-Halo. (d) H4Δ1-19 can be incorporated into both euchromatin and heterochromatic regions. The overall motion of nucleosomes with H4Δ1-19 was lower than that of wtH4-Halo, which was primarily concentrated in the euchromatin.















