More constrained chromatin in oncogenic HRAS-transformed cells.
Maeshima Group / Genome Dynamics Laboratory
Kurokawa Group / Genome Evolution Laboratory
Kuraku Group / Molecular Life History Laboratory
Chromatin organization and behavior in HRAS-transformed mouse fibroblasts
Aoi Otsuka*, Katsuhiko Minami*, Koichi Higashi, Akane Kawaguchi, Sachiko Tamura, Satoru Ide, Michael J. Hendzel, Ken Kurokawa, Kazuhiro Maeshima#
* These authors equally contributed to this work. #corresponding author
Chromosoma 2024 Feb 24 DOI:10.1007/s00412-024-00817-x
Genomic DNA is organized three-dimensionally in the nucleus as chromatin. Recent super-resolution imaging and Hi-C studies have shown that chromatin in living cells forms a number of condensed chromatin domains. Inside these domains, nucleosomes behave locally like a liquid, and such behavior is deeply related to various DNA functions. Cancer cells often show upregulated transcription and other activities, but how is the chromatin behavior in cancer cells different from non-cancer cells?
A research team led by Professor Kazuhiro Maeshima of Genome Dynamics Laboratory (NIG), including a SOKENDAI graduate student Aoi Otsuka (SOKENDAI Special Researcher), SOKENDAI graduate student Katsuhiko Minami (former SOKENDAI Special Researcher, JSPS Research Fellow DC2), Assistant Professor Satoru Ide, Technical Staff Sachiko Tamura, together with Assistant Professor Koichi Higashi and Professor Ken Kurokawa of Genome Evolution Laboratory (NIG), Assistant Professor Akane Kawaguchi of Molecular Life History Laboratory (NIG), and Professor Michael J. Hendzel of University of Alberta, have found chromatin in oncogenic RAS-transformed cells is more constrained than their parental cells.
In this work, the research team examined the chromatin behavior in mouse oncogenic HRAS-transformed cells (CIRAS-3 cells) and their parental cells (10T1/2 cells) with multiple approaches. First, they found that the CIRAS-3 cells have smaller nuclei. In addition, they investigated the individual nucleosome movements in living cells using super-resolution fluorescence microscopy and revealed that nucleosomes are locally more constrained in CIRAS-3 cells. Consistently, CIRAS-3 cells show increased heterochromatin, and in situ Hi-C revealed enriched interactions of the B-B compartments, which mainly consist of heterochromatin.
Genome chromatin in cancer cells encounters physical stress during migration through small spaces, such as metastasis. The smaller nuclei and the more constrained chromatin with enriched heterochromatin can gain elastic properties of chromatin, and may better benefit the cell metastasis (Figure).
This work was supported by JSPS Fellowship, JSPD grants (21H02453, 23K17398, 23K05798, 22H05606, 21H02535, 23KJ0998, 20H05936, 22H04925 (PAGS)), a Japan Science and Technology Agency JST SPRING(JPMJSP2104), The Naito Foundation, and the Takeda Science Foundation. Genome analysis was performed with support of Platform for Advanced Genome Science (22H04925 (PAGS)).
The journal “Chromosoma” is a historical chromosome journal first published in 1939, and is available from the first issue in the NIG Library.
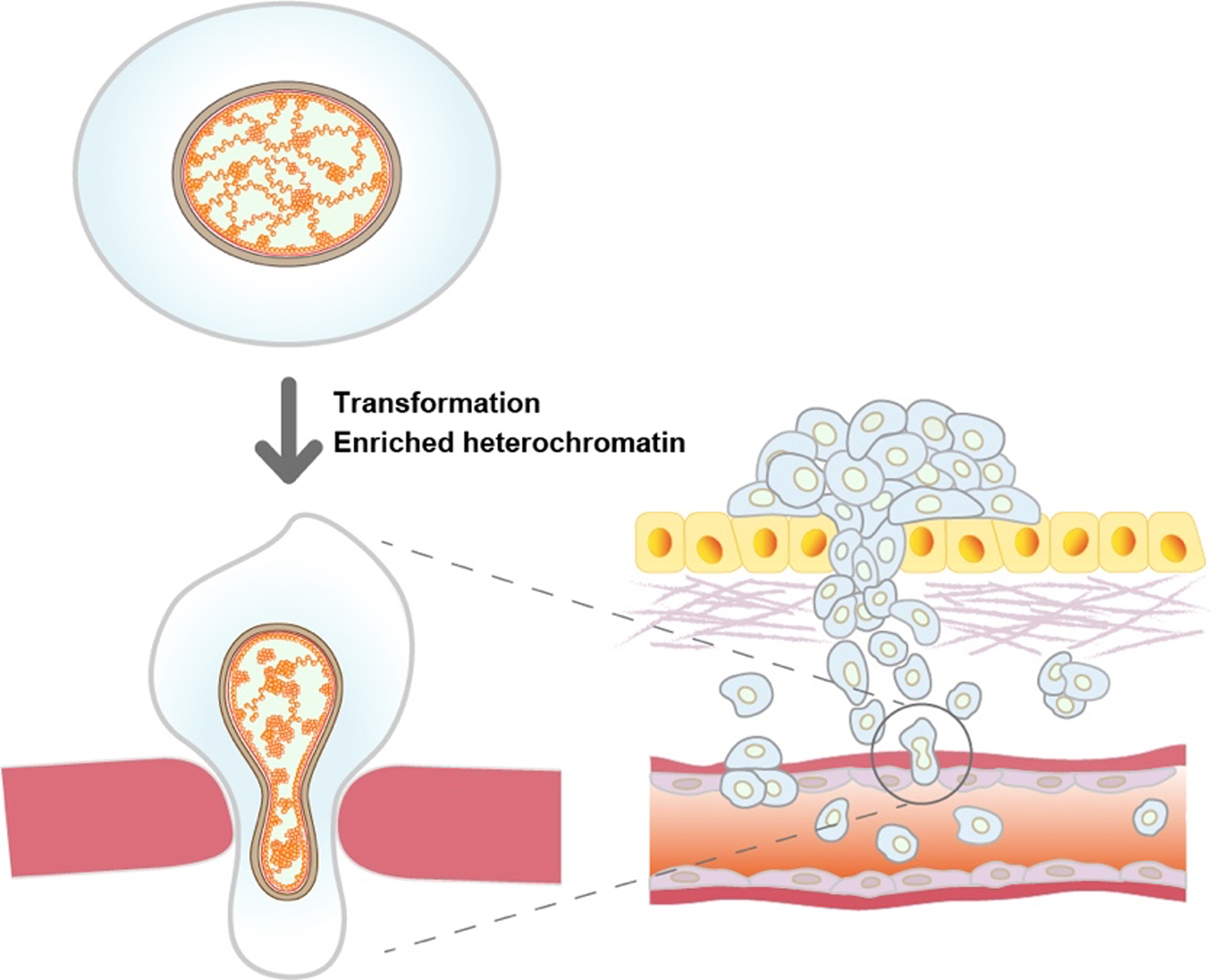
Figure: Oncogenic HRAS-transformed cells (bottom left) have more constrained chromatin with increased heterochromatin than non-transformed parental cells (top left). The more constrained chromatin may play an important role in the metastasis process.















