Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Coordination of cohesin and DNA replication observed with purified proteins
Press release
Coordination of cohesin and DNA replication observed with purified proteins
Yasuto Murayama, Shizuko Endo, Yumiko Kurokawa, Ayako Kurita, Sanae Iwasaki, Hiroyuki Araki
Nature 2024 Jan 24 DOI:10.1038/s41586-023-07003-6
Two newly duplicated copies of genomic DNA are held together by the ring-shaped cohesin complex to ensure faithful inheritance of the genome during cell division. Cohesin mediates sister chromatid cohesion by topologically entrapping two sister DNAs during DNA replication, but how cohesion is established at the replication fork is poorly understood.
Here, we studied the interplay between cohesin and replication by reconstituting a functional replisome using purified proteins. Once DNA is encircled before replication, the cohesin ring accommodates replication in its entirety, from initiation to termination, leading to topological capture of newly synthesized DNA. This suggests that topological cohesin loading is a critical molecular prerequisite to cope with replication. Paradoxically, topological loading per se is highly rate limiting and hardly occurs under the replication-competent physiological salt concentration. This inconsistency is resolved by the replisome-associated cohesion establishment factors Chl1 helicase and Ctf4, which promote cohesin loading specifically during continuing replication. Accordingly, we found that bubble DNA, which mimics the state of DNA unwinding, induces topological cohesin loading and this is further promoted by Chl1.
Thus, we propose that cohesin converts the initial electrostatic DNA-binding mode to a topological embrace when it encounters unwound DNA structures driven by enzymatic activities including replication. Together, our results show how cohesin initially responds to replication, and provide a molecular model for the establishment of sister chromatid cohesion.
Ancient Feces Reveals Intestinal Environment of Hunters-Gatherers, Jomon People
~Metagenomic analysis of human coprolites~
Press release
Metagenomic analyses of 7000 to 5500 years old coprolites excavated from the Torihama shell-mound site in the Japanese archipelago
Luca Nishimura, Akio Tanino, Mayumi Ajimoto, Takafumi Katsumura, Motoyuki Ogawa, Kae Koganebuchi, Daisuke Waku, Masahiko Kumagai, Ryota Sugimoto, Hirofumi Nakaoka, Hiroki Oota, Ituro Inoue
PLOS ONE (2024) 19, e0295924 DOI:10.1371/journal.pone.0295924
![]() Press release (In Japanese only)
Press release (In Japanese only)
The characteristics of the intestinal environment of the Jomon people who lived in the Japanese archipelago thousands of years ago were not known.
In this study, we conducted the first metagenomic analysis in Japan using ancient DNA extracted from four Jomon-period feces fossil (coprolite) samples. Analysis of the large scale DNA data obtained by metagenomic analysis revealed genome sequences derived from bacteria and viruses that are presumed to have existed in the intestines. The results of this analysis may reflect the characteristics of the Jomon people’s intestinal environment.
We plan to conduct similar analyses on other Jomon coprolites to identify more bacteria and viruses, and to elucidate the evolution of intestinal bacteria and viruses from the Jomon period to the present day, as well as the detailed characteristics of the Jomon intestinal environment.
The research was conducted by a joint research group including Luca Nishimura (a graduate student at SOKENDAI) and Project Professor Ituro Inoue from the National Institute of Genetics, Professor Hiroki Oota from Graduate School of Science, The University of Tokyo, and Mayumi Ajimoto, a curator at the Wakasa History Museum in Fukui Prefecture.
The research results were published in the international scientific journal PLOS ONE on January 25, 2024 (JST).
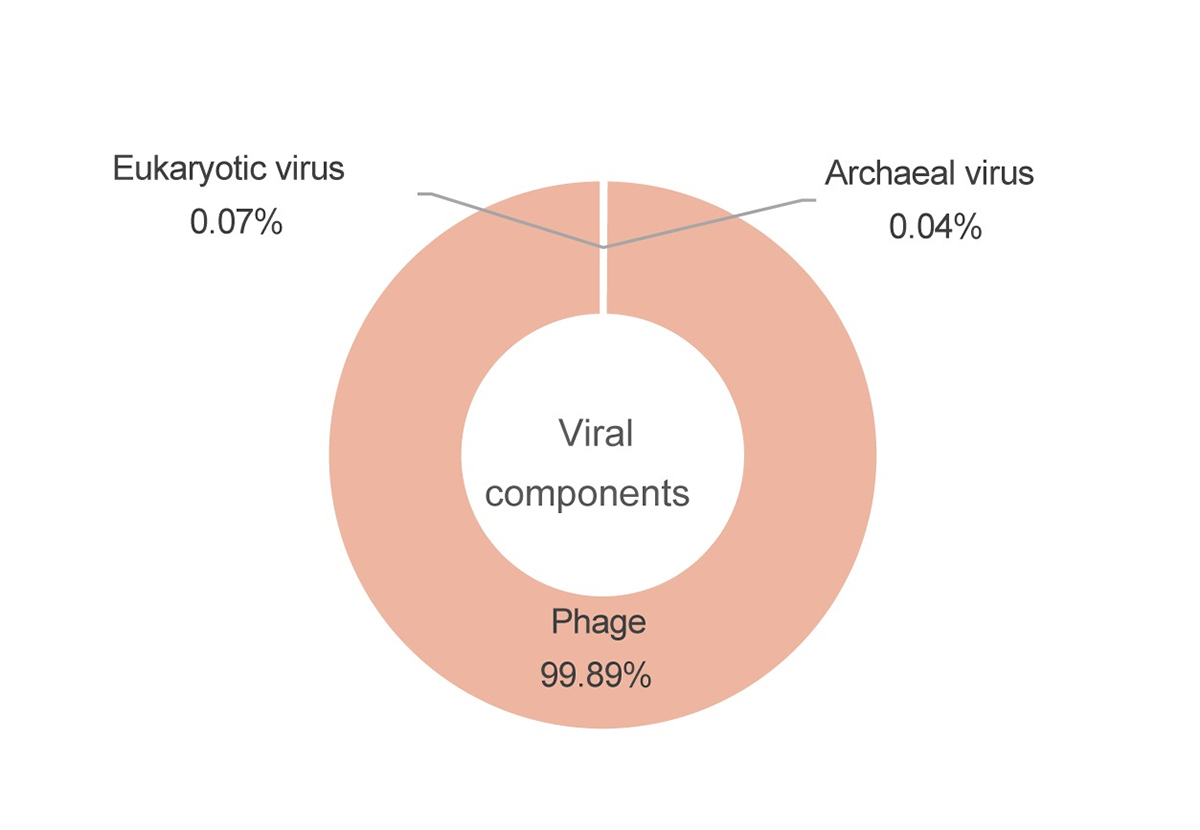
Figure: Classification of viral genome sequences detected in the Jomon coprolites.
The pie chart shows the ratio of virus from eukaryotes as 0.07%, archaea as 0.04%, and viruses infecting bacteria (phage), as 99.89%. You can see that most of the viruses are phages that
Bhim Bahadur Biswa in Mouse Genomics Resource Laboratory won the “MBSJ2023 Best Science Pitch Award” at the MSBJ2023
Bhim Bahadur Biswa(D5 student and SOKENDAI Special Researcher) in Mouse Genomics Resource Laboratory won the “MBSJ2023 Best Science Pitch Award” at the 46th Annual Meeting of the Molecular Biology Society of Japan (MBSJ2023)
- Poster Title: Changes in the gut microbiome associated with domestication of mice

ISLAM Moutushi in Molecular Cell Engineering Laboratory won the “Poster Award” at the SOKENDAI Retreat
ISLAM Moutushi in Molecular Cell Engineering Laboratory won the “Poster Award” at the SOKENDAI Retreat.
- 受賞発表タイトル: Targeted protein degradation with a single-chain antibody-based degron system.

ISLAM Moutushi in Molecular Cell Engineering Laboratory won the “MBSJ2023 Award for EMBO Poster Clinic” at the MSBJ2023
ISLAM Moutushi in Molecular Cell Engineering Laboratory won the “MBSJ2023 Award for EMBO Poster Clinic” at the 46th Annual Meeting of the Molecular Biology Society of Japan (MBSJ2023)
- Poster Title: Targeted protein degradation with a single-chain antibody-based AID2 system

Retina-derived signals control pace of neurogenesis in visual brain areas but not circuit assembly
Kawakami Group / Laboratory of Molecular and Developmental Biology
Retina-derived signals control pace of neurogenesis in visual brain areas but not circuit assembly
Sherman, S., Arnold-Ammer, I., Schneider, M.W., Kawakami, K., and Baier, H.
Nature Communications (2023) 14, 6020 DOI:10.1038/s41467-023-40749-1
Brain development is orchestrated by both innate and experience-dependent mechanisms, but their relative contributions are difficult to disentangle. Here we asked if and how central visual areas are altered in a vertebrate brain depleted of any and all signals from retinal ganglion cells throughout development. We transcriptionally profiled neurons in pretectum, thalamus and other retinorecipient areas of larval zebrafish and searched for changes in lakritz mutants that lack all retinal connections. Although individual genes are dysregulated, the complete set of 77 neuronal types develops in apparently normal proportions, at normal locations, and along normal differentiation trajectories. Strikingly, the cell-cycle exits of proliferating progenitors in these areas are delayed, and a greater fraction of early postmitotic precursors remain uncommitted or are diverted to a pre-glial fate. Optogenetic stimulation targeting groups of neurons normally involved in processing visual information evokes behaviors indistinguishable from wildtype. In conclusion, we show that signals emitted by retinal axons influence the pace of neuro- genesis in visual brain areas, but do not detectably affect the specification or wiring of downstream neurons.
This study was conducted as collaboration with Baier lab at Max Planck Institute.
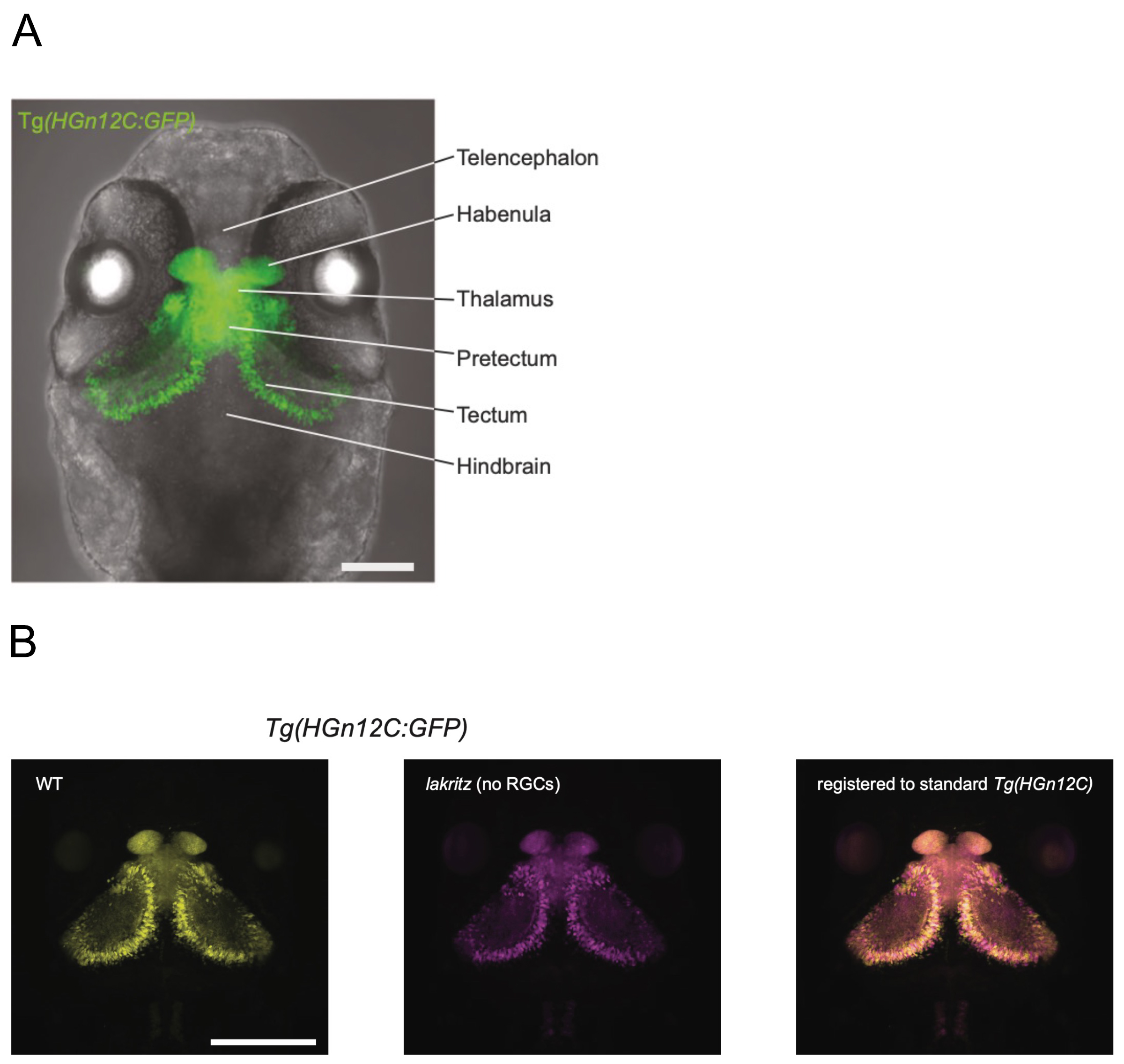
Figure: (A)Transgenic zebrafish (HGn12C) expressing GFP in neurons of retinorecipient areas.
(B)The lakritz mutation was introduced into HGn12C (center), and transcriptional profiling analysis of GFP-positive neurons was conducted.
In vitro storage of functional sperm at room temperature in zebrafish and medaka
Sakai Group / Model Fish Genetics Laboratory
In vitro storage of functional sperm at room temperature in zebrafish and medaka
Takemoto K, Nishimura T, Kawasaki T, Imai Y, Levy K, Hart N, Olaya I, Burgess SM, Elkouby YM, Tanaka M, Sakai N,
Zebrafish (2023) 20, 229-235 DOI:10.1089/zeb.2023.0054
A PDF of this paper can be downloaded from here.
The longevity of sperm in teleost such as zebrafish and medaka is short when isolated even in saline-balanced solution at a physiological temperature. However, we thought that under suitable conditions sperm could be survived for a longer period of time, and we investigated these conditions. Here, we show that the sperm of zebrafish can survive and maintain fertility in L-15-based storage medium supplemented with 50 Units/ml penicillin, 50 µg/ml streptomycin, 0.5% (w/v) bovine serum albumin, 3% foetal bovine serum, 25 mM glucose, 0.1 mM lactic acid, 10 mM Hepes (pH 7.9), and 22% water for 28 days at room temperature. The fertilized embryos developed to normal fertile adults. This storage medium with increasing lactic acid two-fold was effective in medaka sperm stored for 7 days at room temperature. These results suggest that sperm from external fertilizer zebrafish and medaka has the ability to survive for at least 4 weeks and 1 week, respectively, in the body fluid-like medium at a physiological temperature. This sperm storage method allows researchers to ship sperm by low-cost methods and to investigate key factors for motility and fertile ability in those sperm.

Figure: Zebrafish cover featuring zebrafish and medaka sperm
Determinants of motor neuron functional subtypes important for locomotor speed
Kawakami Group/Laboratory of Molecular and Developmental Biology
Determinants of motor neuron functional subtypes important for locomotor speed
Kristen P. D’Elia, Hanna Hameedy, Dena Goldblatt, Paul Frazel, Mercer Kriese, Yunlu Zhu, Kyla R. Hamling, Koichi Kawakami, Shane A. Liddelow, David Schoppik, and Jeremy S. Dasen
Cell Reports (2023) 42, 113049. DOI:10.1016/j.celrep.2023.113049
Locomotion requires precise control of the strength and speed of muscle contraction and is achieved by recruiting functionally distinct subtypes of motor neurons (MNs). MNs are essential to movement and differentially susceptible in disease, but little is known about how MNs acquire functional subtype-specific features during development. Using single-cell RNA profiling in embryonic and larval zebrafish, we identify novel and conserved molecular signatures for MN functional subtypes and identify genes expressed in both early post- mitotic and mature MNs. Assessing MN development in genetic mutants, we define a molecular program essential for MN functional subtype specification. Two evolutionarily conserved transcription factors, Prdm16 and Mecom, are both functional subtype-specific determinants integral for fast MN development. Loss of prdm16 or mecom causes fast MNs to develop transcriptional profiles and innervation similar to slow MNs. These results reveal the molecular diversity of vertebrate axial MNs and demonstrate that functional subtypes are specified through intrinsic transcriptional codes.
This study was conducted as collaboration with Schoppik lab at New York University.
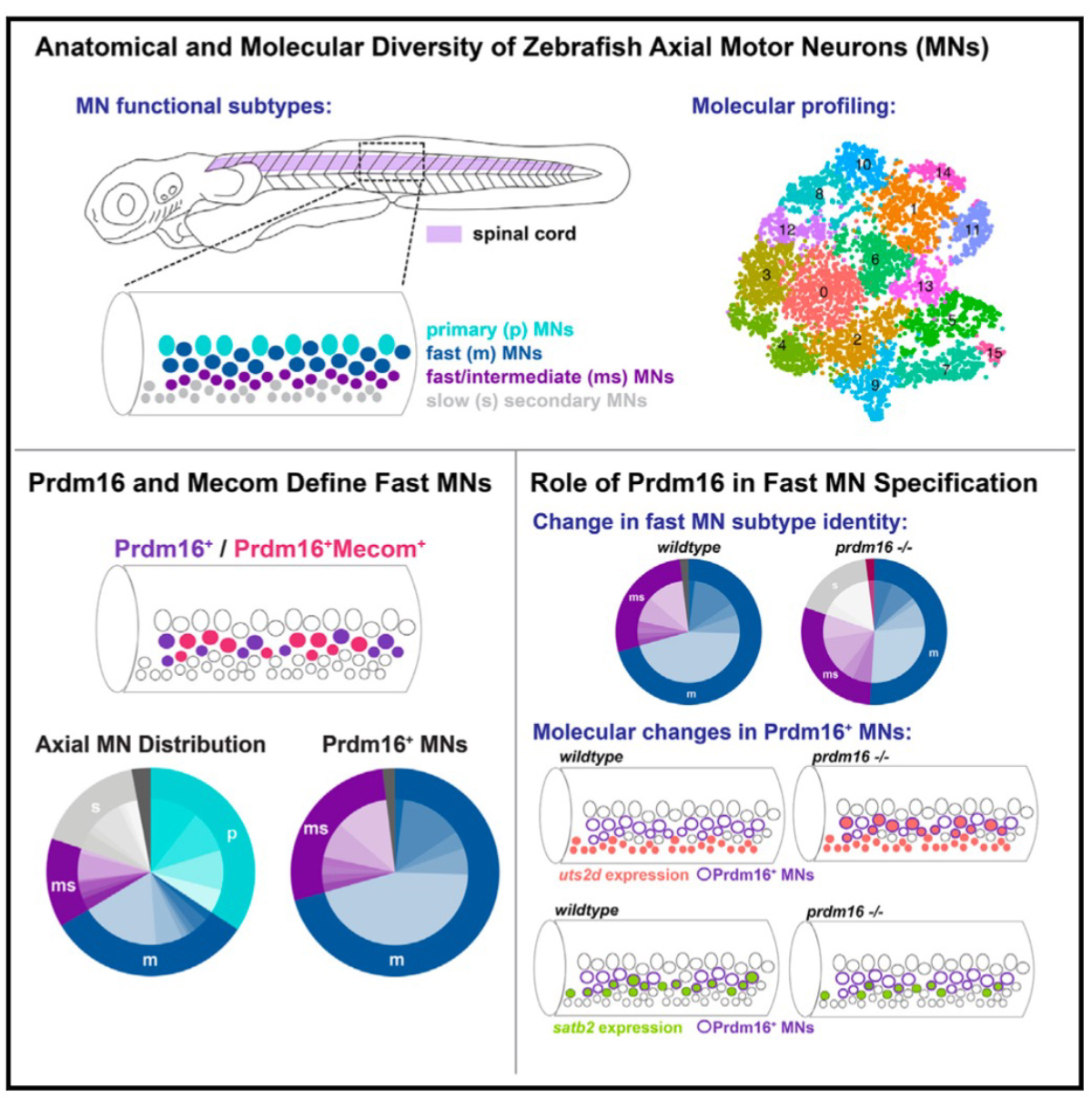
Figure1: Zebrafish motor neuron diversity is regulated by the transcription factors prdm16 and mecom. The absence of these transcription factors results in fast motor neurons exhibiting characteristics of slow motor neurons.
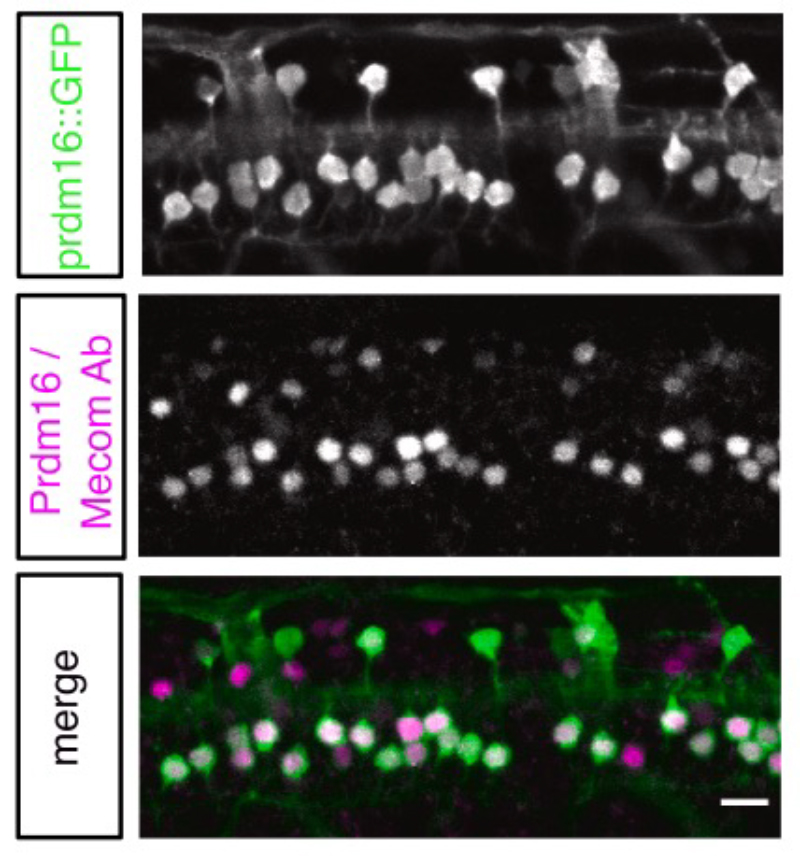
Figure2: Overlap of prdm16 and mecom expression in motor neurons using transgenic fish.
Shiori Iida at Genome Dynamics Laboratory won the “Early Career Presentation Award” and the “IUPAB student award” at the 61st Annual Meeting of the Biophysical Society of Japan
Shiori Iida, a D4 student and JSPS Research Fellow at Genome Dynamics Laboratory, was honored with the “Early Career Presentation Award” and the “IUPAB student award” during the 61st Annual Meeting of the Biophysical Society of Japan in Nagoya, Japan, held from November 14th to 16th. The “IUPAB student award” is granted to a maximum of three outstanding presenters among the students who applied for the Early Career Award / Early Career Presentation Award. Recipients of this award received a certificate and a monetary prize.
- Awarded presentation title: Higher order structure of chromatin regulates local chromatin motion and chromatin stiffness


Left: Ms. Iida holding the certificate
Right: Ms. Iida receiving the certificate from President Takahashi of the Biophysics Society of Japan
Unraveling the evolutionary origins of umami and sweet taste preferences
Press release
A vertebrate-wide catalogue of T1R receptors reveals diversity in taste perception
Hidenori Nishihara, Yasuka Toda, Tae Kuramoto, Kota Kamohara, Azusa Goto, Kyoko Hoshino, Shinji Okada, Shigehiro Kuraku, Masataka Okabe, Yoshiro Ishimaru
Nature Ecology & Evolution 2023 Dec 13 DOI:10.1038/s41559-023-02258-8
![]() Press release (In Japanese only)
Press release (In Japanese only)
EurekAlert! link about this artcle
The perception of taste is one of the most important senses and helps us identify beneficial foods and avoid harmful substances. For instance, our fondness for sweet and savory foods results from our need to consume carbohydrates and proteins. Given their importance as an evolutionary trait, researchers around the world are investigating how taste receptors originated and evolved over a period of time. Obtaining these insights into the feeding behavior of organisms can help them paint a picture of the history of life on Earth. read more>
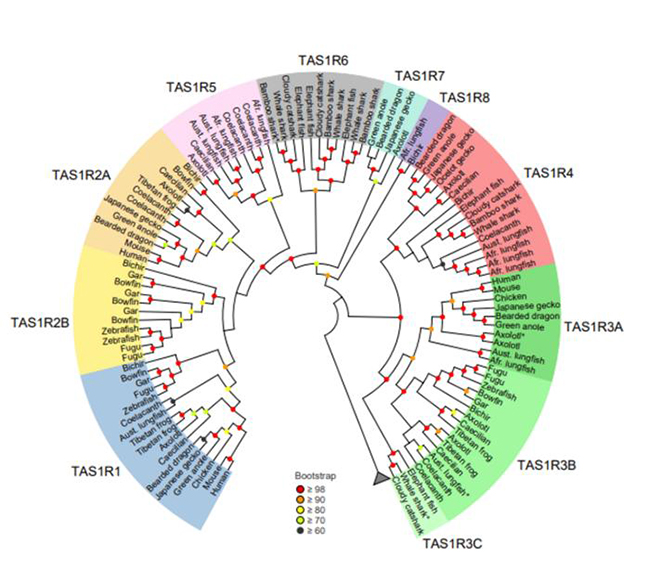
Figure: A new study led by researchers from Kindai University identified five new groups of umami and sweet taste receptors within the TAS1R family (TAS1R 4, 5, 6, 7, and 8) and also diversity in TAS1R2 and TAS1R3 genes.
Call for Urgent Joint Research with Researchers in Noto Peninsula Earthquake Disaster Area
Masa A. Shimazoe in Genome Dynamics Laboratory won the “Student Presentation Award” at the 61st Annual Meeting of the Biophysical Society of Japan
Masa A. Shimazoe (D2, Department of Genetics, SOKENDAI) in Genome Dynamics Laboratory received the “Student Presentation Award” at the 61st Annual Meeting of the Biophysical Society of Japan held in Nagoya, Japan, November 14-16, 2023.
- Title of Presentation: Linker Histone H1 serves as Liquid-like “Glue” for the Chromatin Domain

















