Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Three genes instrumental in increasing harvests in the earlier step of rice domestication
A stepwise route to domesticate rice by controlling seed shattering and panicle shape
Ryo Ishikawa, Cristina Cobo Castillo, Than Myint Htun, Koji Numaguchi, Kazuya Inoue, Yumi Oka, Miki Ogasawara, Shohei Sugiyama, Natsumi Takama, Chhourn Orn, Chizuru Inoue, Ken-Ichi Nonomura, Robin Allaby, Dorian Q Fuller and Takashige Ishii
PNAS (2022) 119, e2121692119 DOI:10.1073/pnas.2121692119
![]() Press release (In Japanese only)
Press release (In Japanese only)
The international collaborative group of Kobe University, National Institute of Genetics (NIG), University of Warwick, University College London, Yezin Agricultural University, Cambodian Agricultural Research and Development Institute, successfully unveiled that the Asian wild rice (O. rufipogon Griff.) dramatically had increased grain yields with mutations at three genes in an initial step of domestication.
The loss of seed-shattering behaviour is an important step of domestication enabling humans to harvest more grains. At first, to experimentally reproduce an earlier process of rice domestication, several domestication-related genes were replaced with those of cultivated rice in the genetic background of wild rice, O. rufipogon. The replacement alone with a cultivated rice-type mutation of sh4, a rice gene with a major impact on seed shattering, was insufficient, but the replacement with cultivated rice-type mutations of both sh4 and qSH3, an allele within the seed-shattering gene OsSh1, caused the partial loss of abscission layer formation in O. rufipogon. However, the combination of two mutations of sh4 and qSH3 was not sufficient to increase yield under natural conditions. To further increase yield, our group found that the replacement together with cultivated rice-type mutations of three genes, sh4, qSH3 and SPR3, dramatically increased yields (Fig. 1). SPR3 is a rice gene controlling a panicle shape, and the closed panicle property enhances long awn-retaining seeds entangled, and in addition, reduces a bending moment which is a predominant factor affecting seed dispersal (Fig. 2), resulting in increased harvests.
We propose a stepwise route in the earliest phase of rice domestication, in which SPR3-controlled closed panicle morphology was instrumental in addition to sequential recruitments of sh4- and qSH3-dependent loss of shattering.
This study was supported partly by NIG-JOINT program 83A2016-2018. The wild rice accession, maintained by NIG in the support of National Bioresource Project (NBRP) Rice, was used in this study.
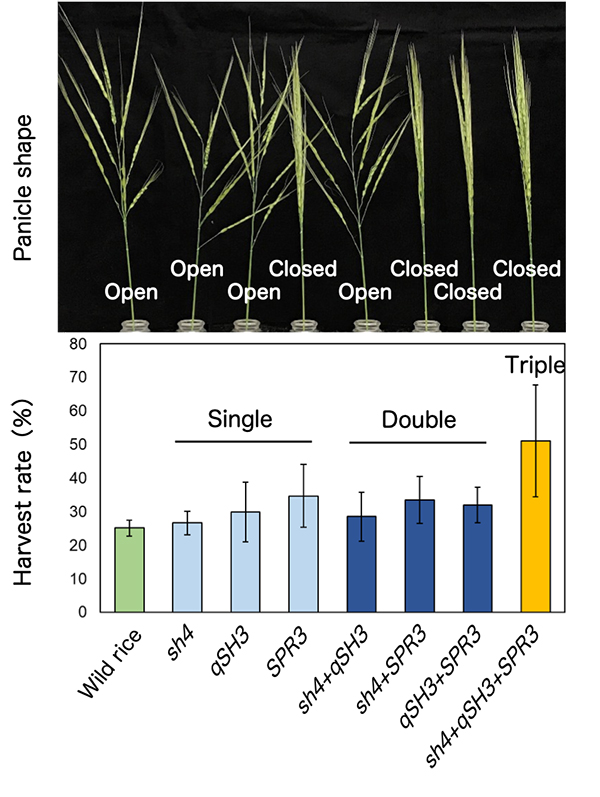
(Top) The panicle shape of seven wild rice introgression lines with different replacements to cultivated rice-type alleles at sh4, qSH3 and SPR3. (bottom) The seed harvest rate of respective wild rice introgression lines.
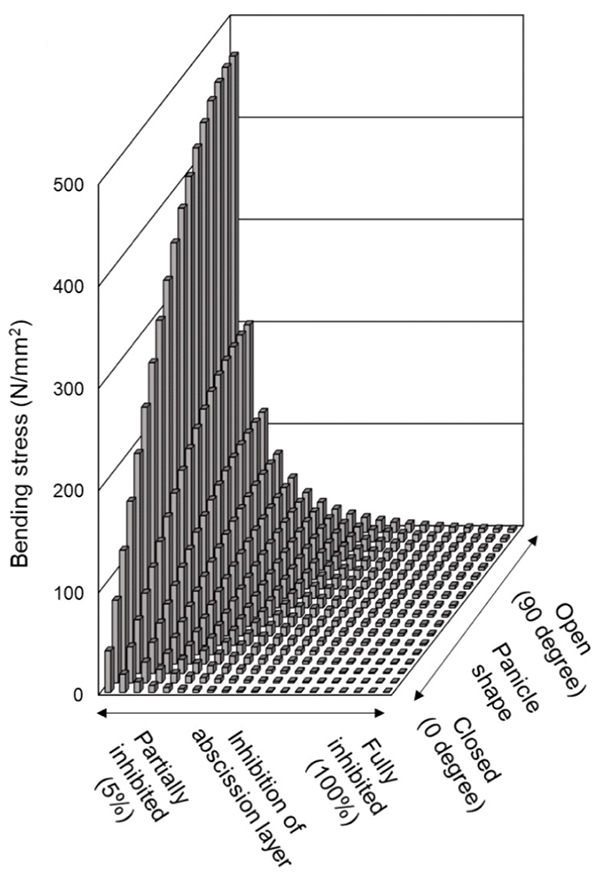
A higher stress was experienced in the open panicle with less inhibited abscission. This simulation suggests that in addiotion to the replacement with sh4 and qSH3 for reduced shuttering, we propose that the replacement with SPR3 for panicles closing might contribute to increased harvest rate during an earlier step of rice domestication.
Detection of Ancient Viruses and Long-Term Viral Evolution
Inoue Group / Human Genetics Laboratory
Detection of Ancient Viruses and Long-Term Viral Evolution
Luca Nishimura, Naoko Fujito, Ryota Sugimoto, and Ituro Inoue*
Viruses (2022) 14, 1336 DOI:10.3390/v14061336
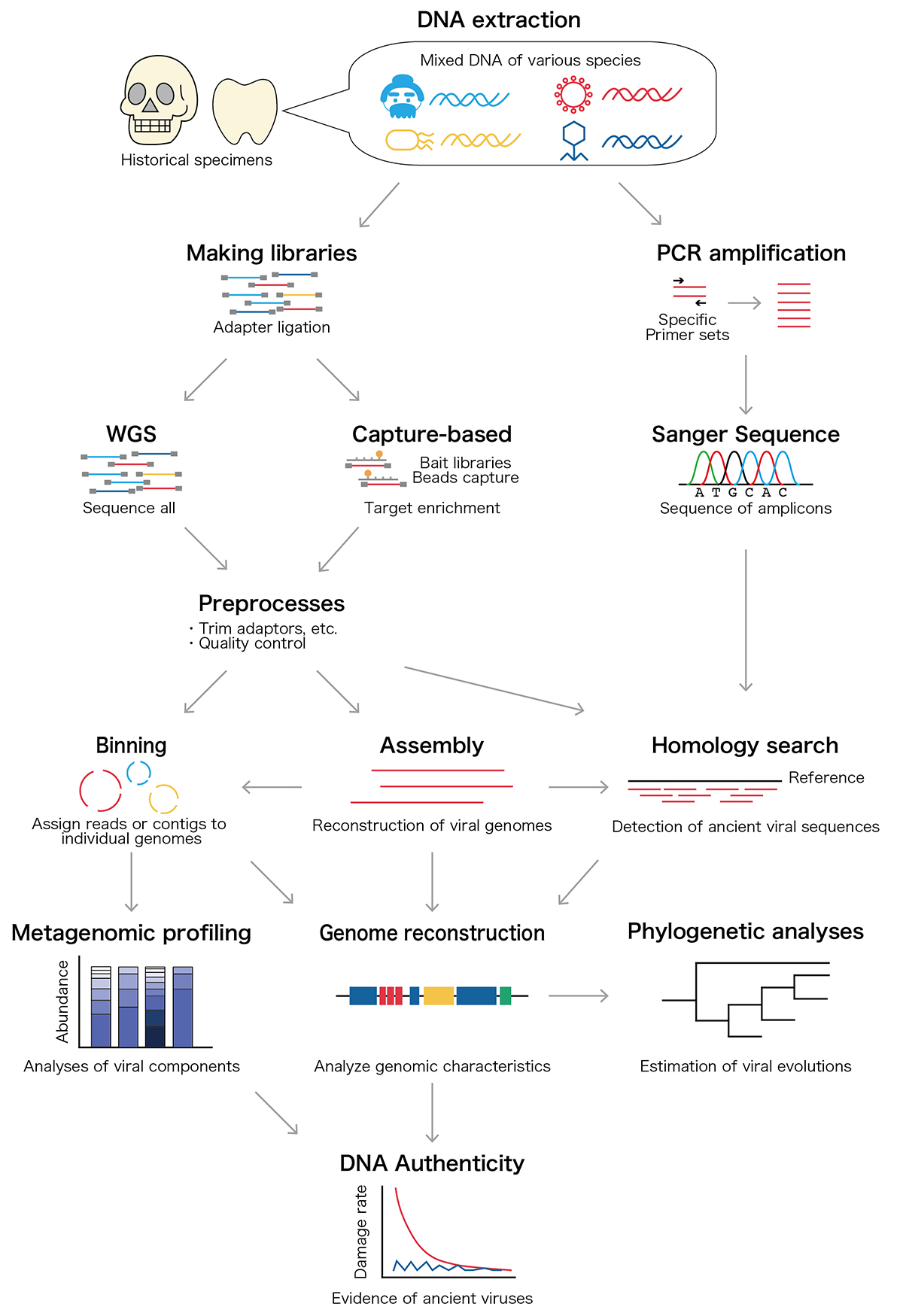
Archaeological remains contain Ancient DNA and RNA, and those nucleic acids provide genomic information about ancient people together with ancient microbiomes and viruses that infected ancient individuals. Since 1997, more than 20 ancient viral species including influenza virus and hepatitis B virus have been discovered from historical samples such as bones and mummified tissues. Those ancient viral genomes have been utilized to estimate the past pandemics of pathogenic viruses within the ancient human population and long-term evolutionary events. In our review article, we overview the ancient viral studies and experimental and analytical techniques and discuss the long-term viral evolutionary studies using ancient viral genomes.
Chromatin behavior in living cells: lessons from single-nucleosome imaging and tracking
Maeshima Group / Genome Dynamics Laboratory
Chromatin behavior in living cells: lessons from single-nucleosome imaging and tracking
*Satoru Ide, Sachiko Tamura, *Kazuhiro Maeshima *corresponding author
BioEssays 2022 June 03 DOI:10.1002/bies.202200043
Eukaryotic genome DNA is wrapped around core histones and forms a nucleosome structure. Together with associated proteins and RNAs, these nucleosomes are organized three-dimensionally in the cell as chromatin. Emerging evidence demonstrates that chromatin consists of rather irregular and variable nucleosome arrangements without the regular fiber structure and that its dynamic behavior plays a critical role in regulating various genome functions. Single-nucleosome imaging is a promising method to investigate chromatin behavior in living cells. It reveals local chromatin motion, which reflects chromatin organization not observed in chemically fixed cells. The motion data is like a gold mine. Data analyses from many aspects bring us more and more information that contributes to better understanding of genome functions. In this review article, we describe imaging of single-nucleosomes and their tracked behavior through oblique illumination microscopy. We also discuss applications of this technique, especially in elucidating nucleolar organization in living cells.
This work was supported by JSPS Grants and MEXT KAKENHI grants (21H02535, 20H05936 and 21H02453) and the Uehara Memorial Foundation.
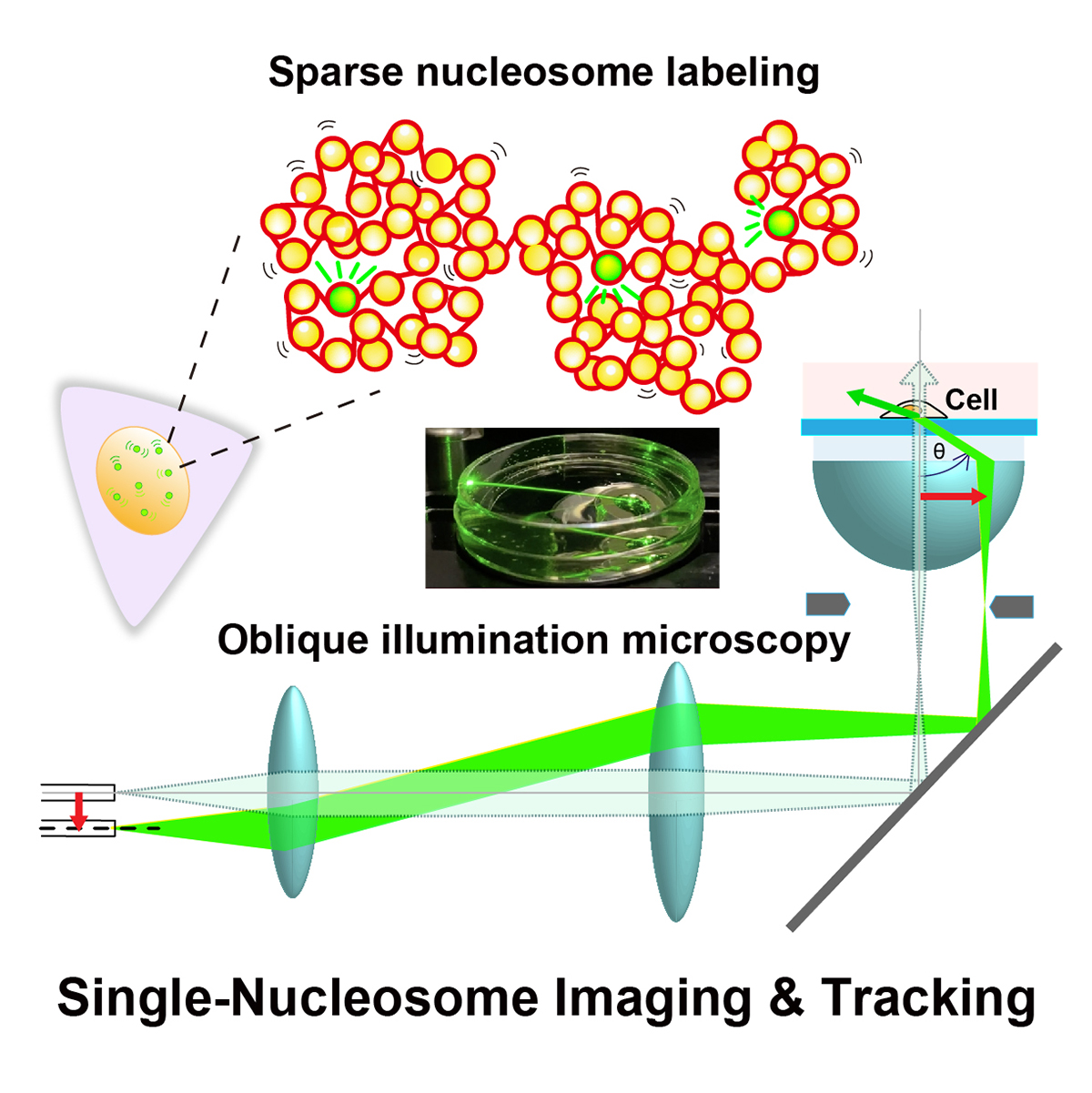
Figure: Single-nucleosome imaging and tracking is a promising technique to investigate dynamic chromatin behavior in living cells. We describe how this imaging works using sparse nucleosome labeling and oblique illumination microscopy, and how information on chromatin dynamics can be extracted from the obtained motion data.
Video1: How laser beams traveled through a glass dish filled with medium using an oblique illumination microscopy. As the incident laser beam shifts from the center axis, higher refraction angles of the beam from the glass are obtained. At the last 2 frames, total internal reflection (TIR) occurs and only the glass surface is illuminated.
Video2: Movie data (50 ms/frame) of single nucleosomes (basic units of chromatin) fluorescently labeled in a living human cell. Note that clear, well-separated dots and their movements were visualized.
Steady-state chromatin motion throughout interphase
Press release
Single-nucleosome imaging reveals steady-state motion of interphase chromatin in living human cells
Shiori Iida, Soya Shinkai, Yuji Itoh, Sachiko Tamura, Masato T. Kanemaki, Shuichi Onami, Kazuhiro Maeshima
Science Advances (2022) 8, eabn5626 DOI:10.1126/sciadv.abn5626
![]() Press release (In Japanese only)
Press release (In Japanese only)
The human body is composed of over forty trillion cells. Each of these cells has totally two meters of tightly packaged genomic DNA, the blueprint of life. Recently, there have been many advances in understanding how DNA is packaged and organized as chromatin in the cell. In contrast, how chromatin behaves in living cells remains unclear.
SOKENDAI graduate student Shiori Iida, JSPS Fellow Yuji Itoh, Technical Stuff Sachiko Tamura, and Professor Kazuhiro Maeshima of Genome Dynamics Laboratory (NIG), together with Research Scientist Soya Shinkai and Team Leader Shuichi Onami of RIKEN BDR, and Professor Masato T. Kanemaki of Molecular Cell Engineering Laboratory (NIG), have investigated the local movements of chromatin in living human cells using super-resolution fluorescence microscopy (Movie 1).
Both DNA amount and nuclear size become double during the preparation period for the cell division (interphase). Previously, it has been suggested that these drastic changes in the nuclear environment would affect chromatin movements. However, Iida et al. have revealed that chromatin motion keeps a steady state throughout the interphase. Chromatin motion is directly related to the accessibility of the DNA (readability of genomic information). Steady-state chromatin motion allows cells to conduct housekeeping tasks under similar environments during interphase.
This work was supported by JSPS and MEXT KAKENHI grants (20H05550、21H05763、19K23735、 20J00572、18H05412、19H05273、20H05936), a Japan Science and Technology Agency CREST grant (JPMJCR15G2), JST SPRING(JPMJSP2104), the Takeda Science Foundation, and the Uehara Memorial Foundation.
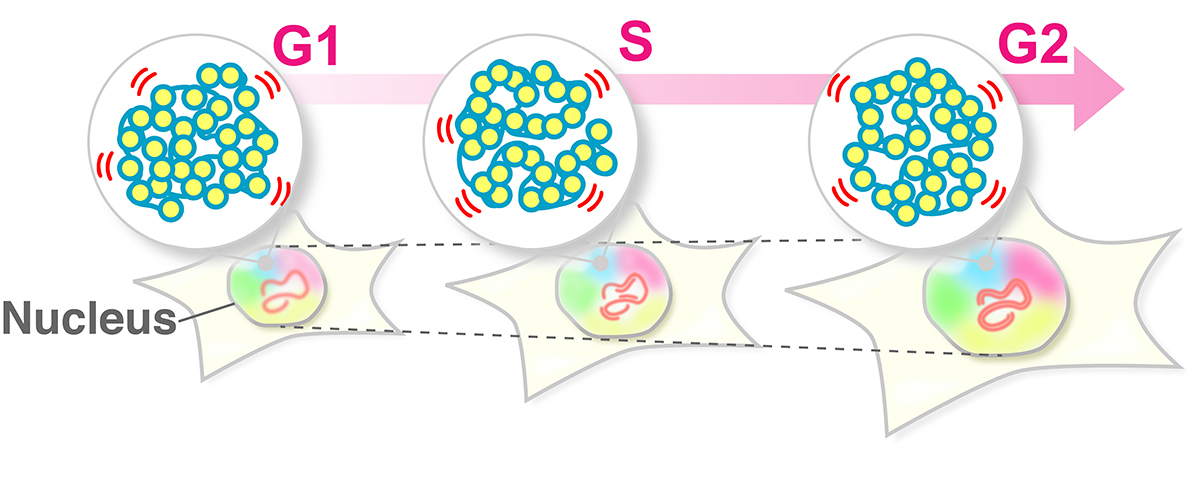
Figure: The swaying motion of chromatin keeps a steady state throughout the interphase (G1, S, G2 phases) despite increases in DNA amount and nuclear volume. Steady-state chromatin motion allows cells to conduct housekeeping tasks under similar environments during interphase.
Video1: Movie data (50 ms/frame) of single nucleosomes (basic units of chromatin) fluorescently labeled in a living human cell. Note that clear, well-separated dots and their movements were visualized.
Video2: Movie data (50 ms/frame) of single nucleosomes fluorescently labeled in living human cells; (left) G1-phase, (right) G2-phase. Note that there is not much difference in nucleosome motion between G1 and G2 phase, even as nuclear size increases.
▶ The article of EurekAlert! is here.















