PZLAST: an ultra-fast amino acid sequence similarity search server against public metagenomes

PZLAST: an ultra-fast amino acid sequence similarity search server against public metagenomes
H. Mori, H. Ishikawa, K. Higashi, Y. Kato, T. Ebisuzaki, K. Kurokawa
Bioinformatics 2021 July 7 DOI:10.1093/bioinformatics/btab492
![]() Press release (In Japanese only)
Press release (In Japanese only)
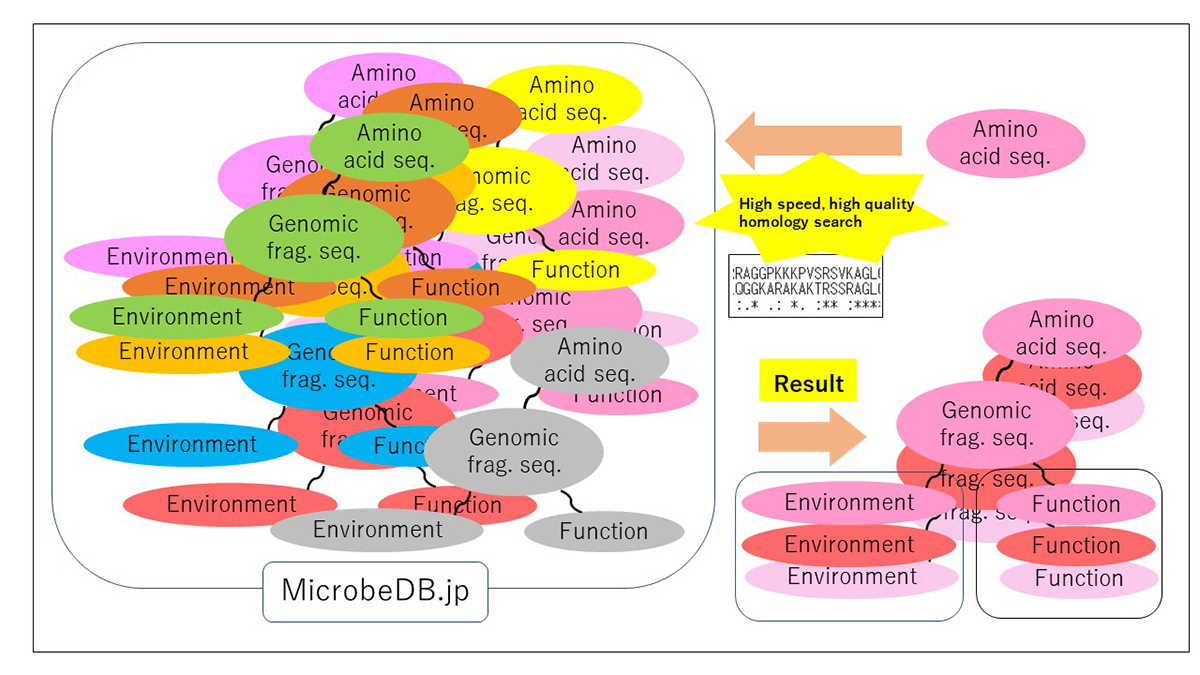
Similarity searches of amino acid sequences against the public metagenomic data can provide users insights about the function of sequences based on the environmental distribution of similar sequences. However, a considerable reduction in the amount of data or the accuracy of the result was necessary to conduct sequence similarity searches against public metagenomic data, because of the vast data size more than Terabytes. Here, we present an ultra-fast service for the highly accurate amino acid sequence similarity search, called PZLAST, which can search the user’s amino acid sequences to several Terabytes of public metagenomic sequences in approximately 10-20 minutes. PZLAST accomplishes its search speed by using PEZY-SC2, which is a MIMD many-core processor. Results of PZLAST are summarized by the ontology-based environmental distribution of similar sequences. PZLAST can be used to predict the function of sequences and mine for homologs of functionally important gene sequences.
Source: H. Mori, et al., Bioinformatics DOI: 10.1093/bioinformatics/btab492
▶ PZLAST is available here.