Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Condensin II makes the chromosome territory in the nucleus
3D genomics across the tree of life reveals condensin II as a determinant of architecture type
Hoencamp, C., Dudchenko, O., Elbatsh, A. M.O., Brahmachari, S., Raaijmakers, J. A., van Schaik, T., Cacciatore, Á. S., Contessoto, V., van Heesbeen, R. G.H.P., van den Broek, B., Mhaskar, A. N., Teunissen, H., St Hilaire, B. G., Weisz, D., Omer, A. D., Pham, M., Colaric, Z., Yang, Z., Rao, S. S.P., Mitra, N., Lui, C., Yao, W., Khan, R., Moroz, L. L., Kohn, A., St. Leger, J., Mena, A., Holcroft, K., Gambetta, M. C., Lim, F., Farley, E., Stein, N., Haddad, A., Chauss, D., Mutlu, A. S., Wang, M. C., Young, N. D., Hildebrandt, E., Cheng, H. H., Knight, C. J., Burnham, T. L.U., Hovel, K. A., Beel, A. J., Mattei, P.-J., Kornberg, R. D., Warren, W. C., Cary, G., Gómez-Skarmeta, J. L., Hinman, V., Lindblad-Toh, K., di Palma, F., Maeshima, K., Multani, A. S., Pathak, S., Nel-Themaat, L., Behringer, R. R., Kaur, P., Medema, R. H., van Steensel, B., de Wit, E., Onuchic, J. N., Di Pierro, M., Lieberman-Aiden, E., Rowland, B. D.
Science 372, 984-989 (2021) DOI:10.1126/science.abe2218
An international collaborative team, which was led by B.D. Rowland at the Netherlands Cancer Institute and E. Lieberman Aiden (NIG International Strategic Advisor) at Baylor College of Medicine, investigated genome folding across the eukaryotic tree of life (24 species, Fig. B). The team found four types of 3D genome architecture at chromosome-scale (Fig. A). Each type appeared and disappeared repeatedly during eukaryotic evolution. The type of genome architecture that an organism exhibits correlates with the absence of condensin II subunits (Fig. B). Condensin is a protein complex required for mitotic chromosome assembly. In vertebrates, it is known that there are two types of condensins, condensin I and II, and that condensin II shortens mitotic chromosomes.
Moreover, the team demonstrated that condensin II depletion converted the architecture of the human genome to a state resembling that seen in organisms such as fungi or mosquitoes (Rabl-like, Fig. C). In this state, centromeres clustered together at nucleoli, and heterochromatin domains merged. The team proposed a physical model in which lengthwise compaction of chromosomes by condensin II during mitosis determines chromosome-scale genome architecture, with effects that are retained during the subsequent interphase. This mechanism is likely conserved since the last common ancestor of all eukaryotes.
The part to which NIG contributed was supported by MEXT KAKENHI grants (20H05936).
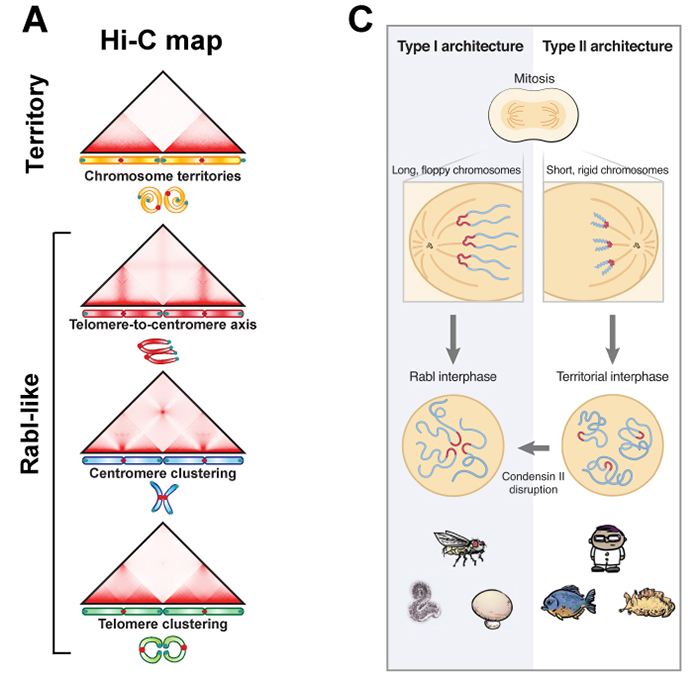
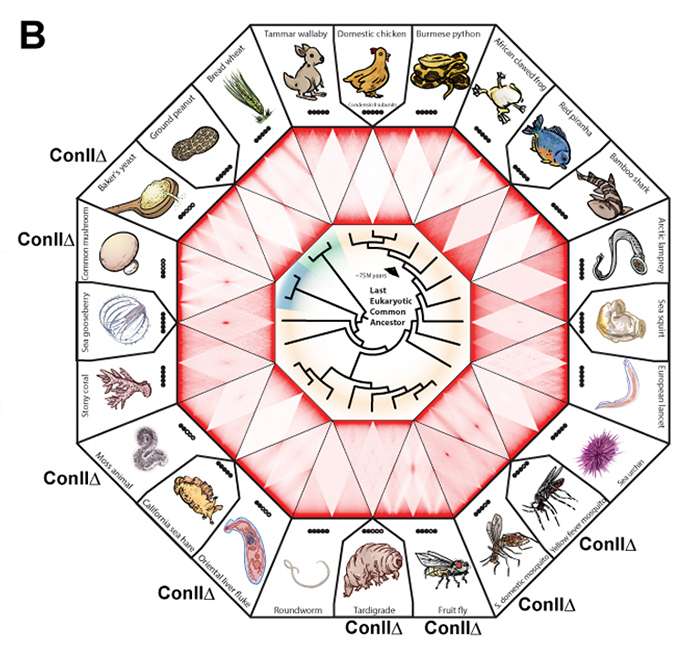
Figure: (A) Chromosome positioning in the nucleus classified by Hi-C maps: “chromosome territory” and “rabl-like”. (B) Hi-C maps of 24 species covering animals (yellow), fungi (blue) and plants (green). Presence of the condensin II subunits in each species is indicated by solid black circles. In case of condensin II-deficiency, shown as “Con IIΔ”. (C) Model. Having shorter chromosomes during cell division leads to separate centromeres and territorial genome architecture in the subsequent interphase. Reducing chromosome lengthwise compaction by condensin II disruption, leads to enhanced centromere clustering, loss of chromosome territories, and a Rabl-like genome architecture.
Notice of the maintenance of NIG website
Our website will be under maintenance from 3:00 p.m. on May 27 to 9:00 a.m. on May 28. The website will be basically available during the maintenance period, but may be intermittently unavailable.
Thank you for your understanding and cooperation.
本日5月27日15:00より翌28日9:00まで、遺伝研ウェブサイトのメンテナンスを予定しています。メンテナンス時間においてもウェブサイトは利用できますが、断続的に利用できない場合があります。
皆様のご理解とご協力をお願いいたします。
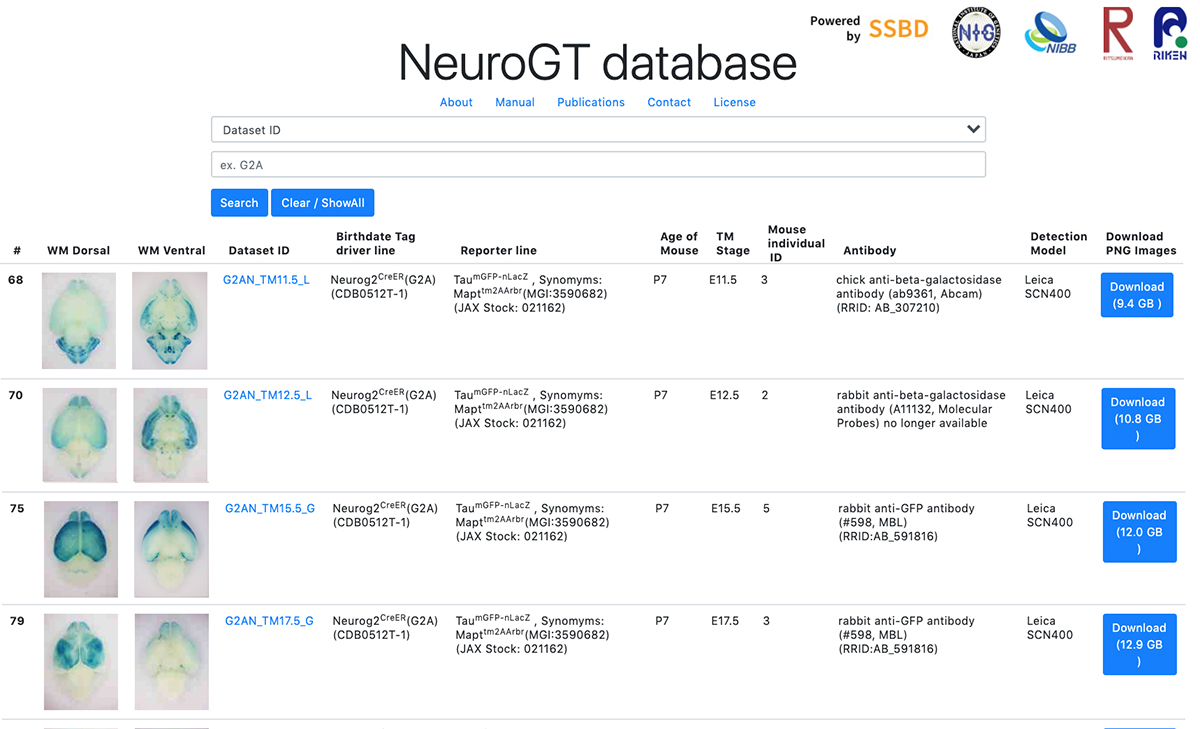
Launch of “NeuroGT Database”, a brain atlas of neurogenic tagging CreER mouse drivers
Press release
NeuroGT: A brain atlas of neurogenic tagging CreER drivers for birthdate-based classification and manipulation of mouse neurons
T Hirata*, Y Tohsato, H Itoga, G Shioi, H Kiyonari, S Oka, T Fujimori, S Onami *Corresponding author
Cell Reports Methods 1, 100012 (2021) DOI:10.1016/j.crmeth.2021.100012
Press release (In Japanese only)
Neuronal birthdate is one of the major determinants of neuronal phenotypes. However, most birthdating methods are retrospective in nature, allowing very little experimental access to the classified neuronal subsets. Hirata and her collaborators have developed four neurogenic tagging mouse lines that can assign CreER-loxP recombination to neuron subsets that share the same differentiation timing in living animals. Because this genetic tag is irreversible, the classified neuronal subset can be subsequently subjected to various experimental manipulations.
To encourage the use of this resource, Hirata et al. have launched “NeuroGT database”, a brain atlas of neurogenic tagging mouse lines, which includes holistic image data of the loxP-recombined neurons and their processes across the entire brain that were tagged at each single day during the neurodevelopmental period. Users can search for the datasets from a web browser using the terns in the meta-information, interactively view thumbnail images of the sections, and download the high-resolution section images.
The NeuroGT is now open to public, offering people the opportunity to find specific neurogenic tagging driver lines and the stages of tagging appropriate for their own research purposes. The driver mouse lines can be obtained from Riken Bioresource Center.
The NeuroGT database was constructed by joint efforts of different research groups; mouse engineering (RIKEN Center for Biosystems Dynamics Research), neuroscience (National Institute of Genetics), bioimaging (National Institute for Basic Biology), and image informatics (Ritsumeikan University, RIKEN Center for Biosystems Dynamics Research)
This research was supported by ROIS Challenging Exploratory Research Projects for the Future Grant and Grant-in-Aid for Publication of Scientific Research Results (Database, 19HP7002). High resolution digitization of section images was supported by Advanced Bioimaging Support (JP16H06280) and Grants-in-Aid for Scientific Research (20H03345, JP18H05412).
- You can use the database here.
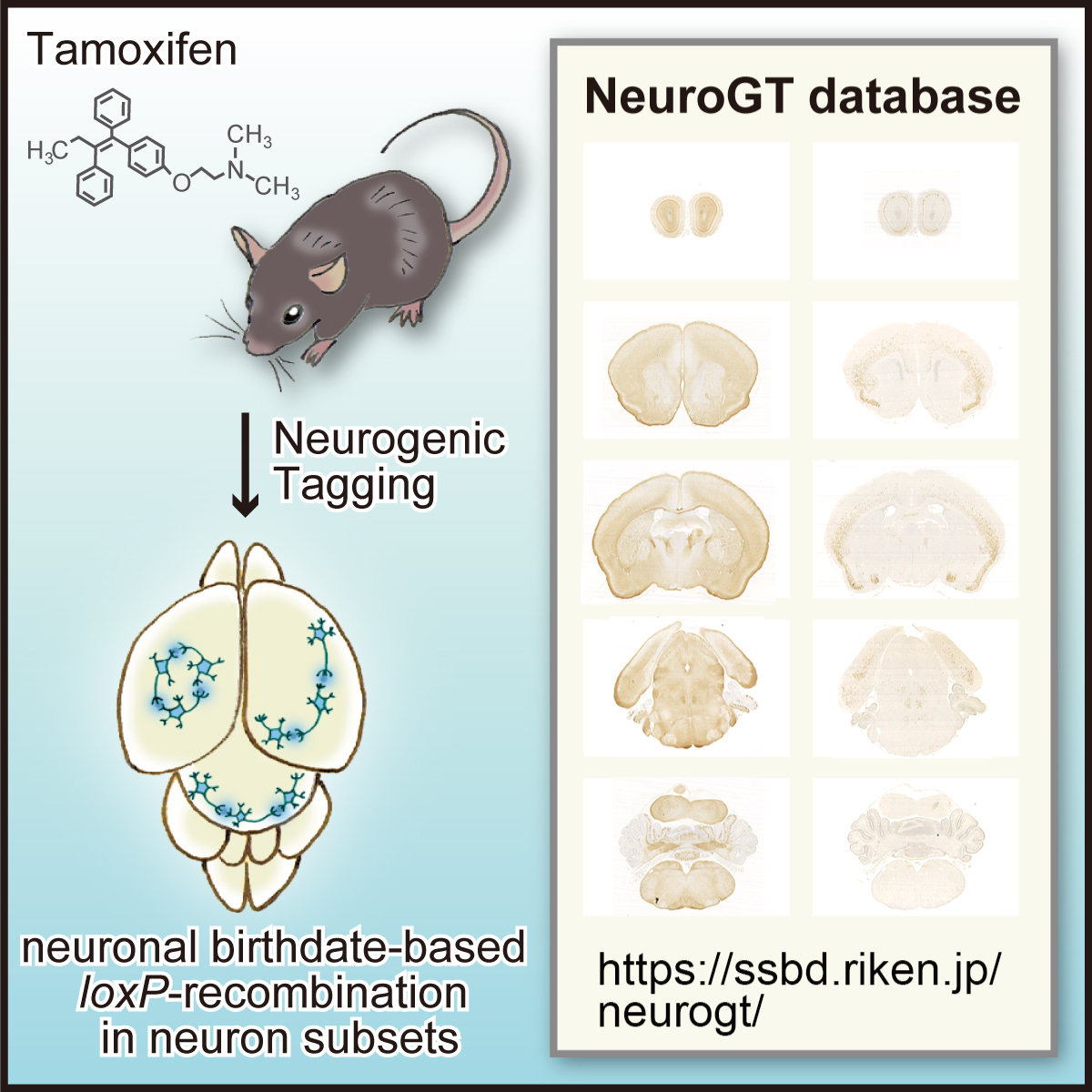
Figure: Graphical abstract of “NeuroGT Database”
Video: Thumbnail images of coronal sections can be sequentially viewed along the antero-posterior brain axis by dragging the slider. The images stained for the two different reporters, membrane-localized GFP and nucleus-localized-βGAL are stacked separately. The sync button automatically matches the section level of the two reporter images.
Signing of a Memorandum of Cooperation with Shizuoka Prefecture for the Implementation of Whole Genome Analysis of Novel Coronavirus.
National Institute of Genetics has offered cooperation to Shizuoka prefecture and has signed a contract concerning investigation cooperation since April 2020. In response to the spread of infection of the new coronavirus variant in these days, a memorandum of cooperation on molecular epidemiological investigation (SARS-CoV-2 RNA whole genome analysis) of novel coronavirus has been signed between our institute and Heita Kawakatsu, Governor of Shizuoka Prefecture, at the government office.
The principles set in the memorandum are: “to support for proactive epidemiological investigation (SARS-CoV-2 RNA whole genome analysis) promoted by Shizuoka prefecture”, “to contribute to treatment of patients infected with novel coronavirus and prevention of the spread of the virus”, “to disclose molecular epidemiological information obtained from specimen samples in order to contribute to the conquest of COVID-19” and “to give due consideration to the protection of personal information when molecular epidemiological information are disclosed”. Based on this memorandum, our institute implements support for SARS-CoV-2 RNA whole genome analysis in earnest.
April 30th, 2021
National Institute of Genetics
Left, Heita Kawakatsu (Governor, Shizuoka Prefecture): Right, Fumio Hanaoka (Director-General, National Institute of Genetics)
Analysis of gene expression profile of pre-granulosa cells during primordial follicle formation
Decoding the transcriptome of pre-granulosa cells during the formation of primordial follicles in the mouse
Kurumi Fukuda, Masafumi Muraoka, Yuzuru Kato, and Yumiko Saga
Biology of Reproduction 2021 April 13 DOI:10.1093/biolre/ioab065
6. Primordial follicles, a finite reservoir of eggs in mammalian ovaries, are composed of a single oocyte and its supporting somatic cells, termed granulosa cells. Although their formation may require reciprocal interplay between oocytes and pre-granulosa cells, precursors of granulosa cells, little is known about the underlying mechanisms. We addressed this issue by decoding the transcriptome of pre-granulosa cells during the formation of primordial follicles. We found that marked gene expression changes, including extracellular matrix, cell adhesion, and several signaling pathways, occur along with primordial follicle formation. Importantly, differentiation of pre-granulosa cells to granulosa cells was delayed in mutant ovaries of the germ cell-specific genes Nanos3 and Figla, accompanied by perturbed gene expression in mutant pre-granulosa cells. These results suggest that proper development of oocytes is required for the differentiation of pre-granulosa cells.
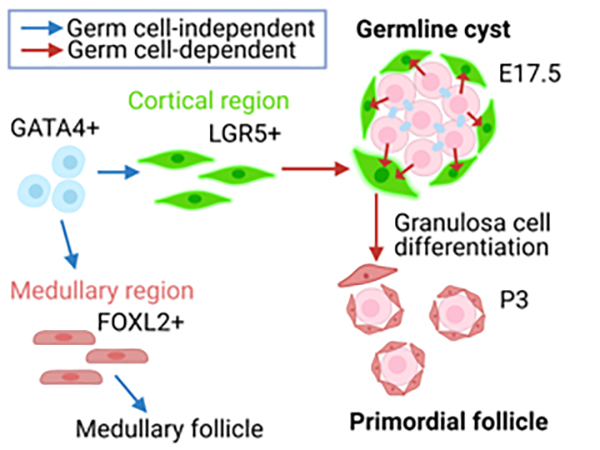
Figure: Pre-granulosa cells (Lgr5+) originate from GATA4 + precursors (blue arrow) in an oocyte-independent manner. On the other hand, differentiation of pre-granulosa cells to granulosa cells requires oocytes (red arrow).















