Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
HLA-B*39:01:01 is a novel risk factor for antithyroid drug-induced agranulocytosis
HLA-B*39:01:01 is a novel risk factor for antithyroid drug-induced agranulocytosis in Japanese population
Saya Nakakura, Kazuyoshi Hosomichi, Shinya Uchino, Akiko Murakami, Akira Oka, Ituro Inoue, Hirofumi Nakaoka
The Pharmacogenomics Journal 2020 September 22 DOI:10.1038/s41397-020-00187-4
Anti-thyroid drug (ATD) is a mainstay of Graves’ disease. About 0.1-0.5% of patients with Graves’ disease treated with ATD exhibit agranulocytosis, which is characterized by severe reduction of circulating neutrophils. Although it has been reported that the HLA class II allele (HLA-DRB1*08:03) was associated with ATD-induced agranulocytosis, the entire HLA region have not been explored in Japanese. Therefore, we performed HLA sequencing for 10 class I and 11 class II genes in 87 patients with ATD-induced agranulocytosis and 384 patients with Graves’ disease who did not develop ATD-induced agranulocytosis. By conducting case-control association studies at the HLA allele and haplotype levels, we identified HLA-B*39:01:01 as an independent risk factor. To verify the reproducibility of the association of HLA-B*39:01:01, we retrieved allele frequency data for HLA-B*39:01:01 from previous case-control association studies. The association of HLA-B*39:01:01 was significantly replicated in Chinese, Taiwanese, and European populations. A meta-analysis combining results from the previous and current studies reinforced evidence of association between HLA-B*39:01:01 and ATD-induced agranulocytosis. The results of this study will provide a better understanding of the pathogenesis of ATD-induced agranulocytosis in the context of HLA-mediated hypersensitivity reaction.
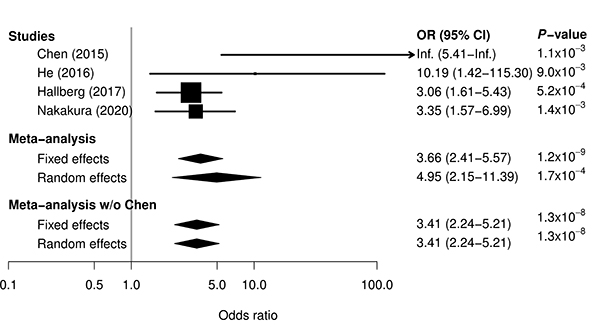
Figure: Meta-analysis for association between ATD-induced agranulocytosis and HLA-B*39:01:01 by combining previous three studies and current study. For single studies, odds ratio and 95% confidence interval are represented by box and whisker. For meta-analyses, odds ratio and 95% confidence interval are represented by a diamond. In all the single studies, HLA-B*39:01:01 was significantly associated with the risk for ATD-induced agranulocytosis. A meta-analysis combining results from the previous and current studies reinforced evidence of the association.
Phase Transitions of Spontaneous Activity Spatial Patterns in Developing Mouse Barrel Cortex
Developmental Phase Transitions in Spatial Organization of Spontaneous Activity in Postnatal Barrel Cortex Layer 4
Shingo Nakazawa, Yumiko Yoshimura, Masahiro Takagi, Hidenobu Mizuno, Takuji Iwasato.
Journal of Neuroscience 2020 September 4 DOI:10.1523/JNEUROSCI.1116-20.2020
Developing sensory cortices exhibit spatially-organized spontaneous activity, which is critical for the cortical circuit maturation. We previously reported “patchwork-type” spatial organization of spontaneous activity in the mouse barrel cortex at postnatal day 5 (P5) (Press release at 2018)
In the present study, we analyzed in detail how the spatial organization of barrel cortex spontaneous activity changes during the first two postnatal weeks. We found that spontaneous activity between P1 to P5 exhibited a patchwork-type pattern (Phase I). While around P9, a new type of pattern, showing wide area synchronization (Phase II), was observed, and at P11, neurons fired sparsely as observed in the adult brain (Phase III).
When thalamus was genetically silenced, Phase I cortical activity was abolished but Phase II and III activity remained intact, suggesting that the Phase I to II transition is associated with the shift of the activity source. On the other hand, the Phase II to III transition was impaired by cortical Rac1 inhibition. Phase II to III transition may be facilitated by Rac1-mediated synapse maturation.
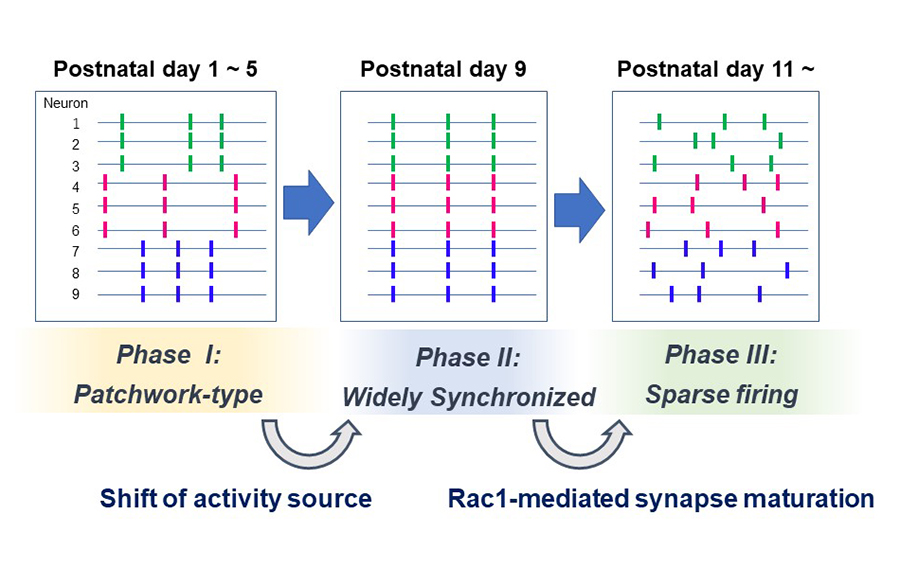
Figure: Three phases of spontaneous network activity were found in the barrel cortex layer 4 during the first two postnatal weeks. The Phase I to II transition is associated with the loss of thalamocortical input dependency. The Phase II to III transition may rely on cortical Rac1-dependent synapse maturation.
Zebrafish can regenerate endoskeleton in larval pectoral fin but the regenerative ability declines
Zebrafish can regenerate endoskeleton in larval pectoral fin but the regenerative ability declines
Keigo Yoshida, Koichi Kawakami, Gembu Abe, Koji Tamura
Developmental Biology 463, 110-123 (2020). DOI:10.1016/j.ydbio.2020.04.010
We show for the first time endoskeletal regeneration in the developing pectoral fin of zebrafish. The developing pectoral fin contains an aggregation plate of differentiated chondrocytes (endochondral disc; primordium for endoskeletal components, proximal radials). The endochondral disc can be regenerated after amputation in the middle of the disc. The regenerated disc sufficiently forms endoskeletal patterns. Early in the process of regenerating the endochondral disc, epithelium with apical ectodermal ridge (AER) marker expression rapidly covers the amputation plane, and mesenchymal cells start to actively proliferate. Taken together with re-expression of a blastema marker gene, msxb, and other developmental genes, it is likely that regeneration of the endochondral disc recaptures fin development as epimorphic limb regeneration does. The ability of endoskeletal regeneration declines during larval growth, and adult zebrafish eventually lose the ability to regenerate endoskeletal components such that amputated endoskeletons become enlarged. Endoskeletal regeneration in the zebrafish pectoral fin will serve as a new model system for successful appendage regeneration in mammals.
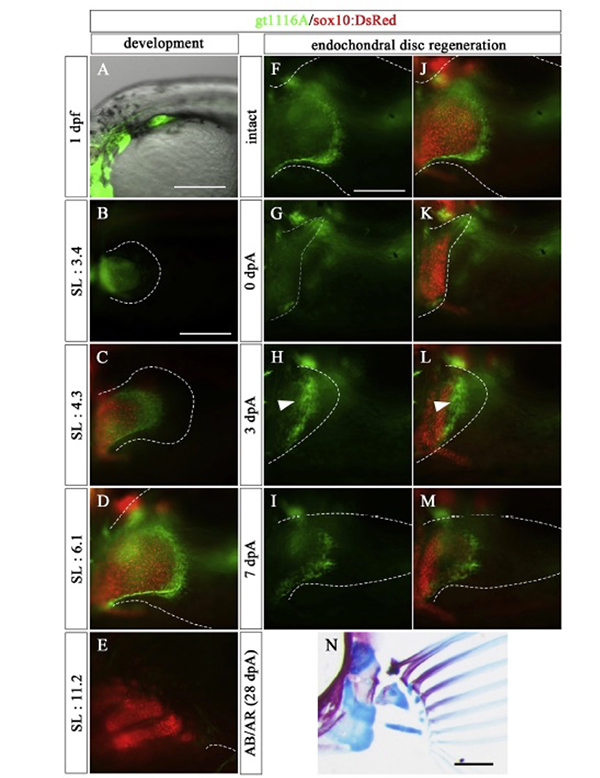
Figure: In the prdm16 gene trap line, the mesenchymal cells of the endoskeletal part of the pectoral fin expressed GFP (green) during the embryonic development. In this line, GFP was also expressed in the mesenchymal cells of the endoskeletal part during endochondrial disc regeneration. The red color in the figure indicates the expression of the sox10 gene, and labeled chondrocytes.
The dynamics, causes and impacts of mammalian evolutionary rates revealed by the analyses of capybara draft genome sequences
Press release
The dynamics, causes and impacts of mammalian evolutionary rates revealed by the analyses of capybara draft genome sequences
Isaac Adeyemi Babarinde and Naruya Saitou
Genome Biology and Evolution (2020) evaa157 DOI:10.1093/gbe/evaa157
Capybara (Hydrochoerus hydrochaeri) is the largest species among the extant rodents. The draft genome of capybara was sequenced with the estimated genome size of 2.6 Gbp. Although capybara is about 60 times larger than guinea pig, comparative analyses revealed that the neutral evolutionary rates of the two species were not substantially different. However, analyses of 39 mammalian genomes revealed very heterogeneous evolutionary rates. The highest evolutionary rate, 8.5 times higher than the human rate, was found in the Cricetidae-Muridae common ancestor after the divergence of Spalacidae. Muridae, the family with the highest number of species among mammals, emerged after the rate acceleration. Factors responsible for the evolutionary rate heterogeneity were investigated through correlations between the evolutionary rate and longevity, gestation length, litter frequency, litter size, body weight, generation interval, age at maturity, and taxonomic order. The regression analysis of these factors showed that the model with three factors (taxonomic order, generation interval and litter size) had the highest predictive power (R2 = 0. 74). These three factors determine the number of meiosis per unit time. We also conducted transcriptome analysis, and found that the evolutionary rate dynamics affects the evolution of gene expression patterns.

Left Panel: The memory of capybara “Rai-chan” (Photo: Izu Shaboten Zoo)
right Panel:Dr. Babarinde (from Nigeria, Africa), who received his Ph.D. from the Graduate University for Advanced Studies, SOKENDAI, in Saito’s lab and is currently a post-doctoral fellow in China.















