Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
The cohesin ring tethers DNA strands
![]()
Establishment of DNA-DNA Interactions by the Cohesin Ring
Yasuto Murayama, Catarina P. Samora, Yumiko Kurokawa, Hiroshi Iwasaki and Frank Uhlmann
CellPublished Online January 18, 2018 DOI:10.1016/j.cell.2017.12.021
Pressrelease (In Japanese only)
DNA, which encodes a blueprint of life, is a very long molecular thread and is packed into a cellular nuclear by forming the protein-DNA supramolecular complex called chromosome. During cell proliferation, chromosomes are doubled up by copying DNA molecules before equal segregation to two daughter cells. To achieve this process smoothly without any mistakes, chromosome contains several special molecular structures. One of such essential chromosomal structures is sister chromatid cohesion. Literally, this is a physical connection which is formed between two duplicated chromosomes. Without cohesion, cell fails to segregate chromosomes properly and this can result in plethora of abnormal events including cell death and cancers. Cohesin is the ring-shaped protein complex and is vital for establishment of sister chromatid cohesion. How cohesin achieves cohesion establishment is one of open questions in chromosome biology.
In the current study, Murayama and colleagues purified the fission yeast cohesin and reconstituted its DNA binding reaction in a test tube. This revealed cohesin topologically entraps DNA inside of its ring. Moreover, cohesin tethers two DNA strands by topological embrace. If DNA-cohesin-DNA tethering is formed between two replicated chromosomes, this can establish chromosome cohesions.
Cohesin is a member of Structural Maintenance of Chromosome (SMC) protein complex family. SMC complexes play vital roles in chromosome organizations. Deficiencies in the complexes have been reported to cause human diseases and malfunctions including developmental disorders, cancers and infertility. The current study provides the first molecular glimpse at how an SMC complex, cohesin, tethers DNA strands.
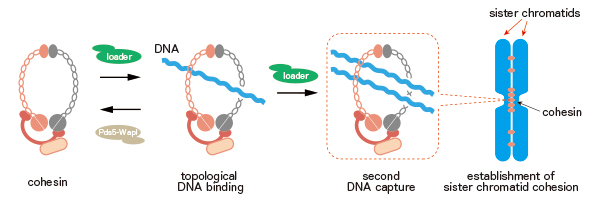
Figure: a model of establishment of sister chromatid cohesion by the cohesin ring.
Cohesin tethers two DNA strands by topological embrace. DNA-cohesin-DNA tethering can provide a molecular bridge to establish cohesion between the replicated sister chromatids.
Assistant Professor Kazuhide Asakawa, Division of Molecular and Developmental Biology won the 1st “SERIKA FUND” award

Assistant Professor Kazuhide Asakawa, Division of Molecular and Developmental Biology won the 1st “SERIKA FUND” award established for ALS (Amyotrophic Lateral Sclerosis) research.
This award is given to scientists who are expected for their innovative and developing research.
The fund is named after heroin of Japanese comic “Space Brothers” who lost her father suffering from ALS. In the story, the heroin Serika becomes an astronaut and succeeds gravity- free-space experiments to develop medicines for ALS treatments. It is hoped to make her wish come true in real life.
▶ Division of Molecular and Developmental Biology
New assistant professor joins NIG
New assistant professor joins NIG as of January 1, 2018.
Keita MIYOSHI: Invertebrate Genetics Laboratory, Saito Group
Release of an upgraded NIG-Mouse Genome Database, “NIG_MoG2”, to the public
Mammalian Genetics Laboratory / Shiroishi Group
Genetic Informatics Laboratory / Kawamoto Group
Commonly used laboratory mouse strains have mosaic genomes, which are derived predominantly from the west European subspecies with the remaining sequences derived from the Japanese subspecies. National Institute of Genetics (NIG) has established a series of experimental mouse strains named “Mishima Battery”. It consists of ten inbred strains originated from four mouse subspecies including the Japanese subspecies, which are widely distributed in the world (Fig. 1). Most of these strains have very remote genetic status from the commonly used laboratory mouse strains such as C57BL/6J (B6), and show very unique phenotypes. Genomic polymorphisms between the laboratory strains and the Japanese subspecies-derived strains are widely used in studies of epigenetics as allele-specific markers. The mouse strains in the “Mishima Battery” are now distributed to the research community via RIKEN BioResource center and NIG, and contribute to broad range of life science. NIG has also established B6-ChrNMSM consomic strains, in which every chromosome of B6 is substituted by corresponding chromosome of Japanese wild mouse-derived MSM/Ms strain, a member of “Mishima Battery”. Using a full panel of the consomic strains, researchers are able to identify chromosomes that control complex traits and diseases. NIG provides these consomic strains to the research community.
Our recent genome-resequencing project of “Mishima Battery” revealed over forty millions of SNPs due to large inter-subspecific genetic distance. Here, we release an upgraded version of the NIG Mouse Genome database named “NIG_MoG2” (http://molossinus.lab.nig.ac.jp/mog2/). NIG_MoG2 primarily comprises the whole genome sequence data of the “Mishima Battery” (Fig. 2), providing visualized genome polymorphism information with single-nucleotide polymorphisms and short insertions/deletions in the genomes of the “Mishima Battery”. It allows users, especially for wet-lab biologists, to intuitively recognize inter-subspecific genome divergence on the browser. NIG_MoG2 is thus a valuable navigator for exploring mouse genome polymorphisms.
This project was supported by NIG Advanced Genomics Project (Next Generation Genome-Research-Hub Project), “Genome Information Upgrading Program” of National BioResource Project (NBRP) from Japan Agency for Medical Research and Development (AMED). The project was performed as a mission of the Genetic Resource Center of NIG.
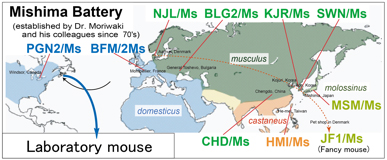
Fig. 1. Geographic distribution of mouse subspecificies, and origins of the “Mishima Battery”. Names of the strains are indicated in colored and bold prints.
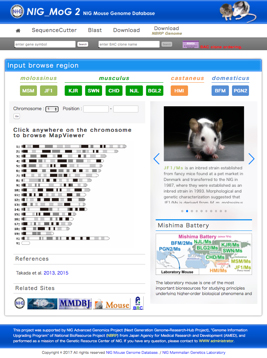
Fig. 2. Top page of the NIG_MoG2 (http://molossinus.lab.nig.ac.jp/mog2).
Patchwork-type spontaneous activity in neonatal barrel cortex layer 4 transmitted via thalamocortical projections
![]()
Patchwork-type spontaneous activity in neonatal barrel cortex layer 4 transmitted via thalamocortical projections
Hidenobu Mizuno, Koji Ikezoe, Shingo Nakazawa, Takuya Sato, Kazuo Kitamura, Takuji Iwasato
Cell Reports Volume 22, Issue 1, p123–135, 2 January 2018 DOI:10.1016/j.celrep.2017.12.012
Pressrelease(In Japanese only)
Establishment of precise neuronal connectivity in the mammalian neocortex relies on activity-dependent circuit reorganization during postnatal development; however, the nature of cortical activity during this period remains largely unknown.
Using two-photon calcium imaging of the mouse somatosensory cortex (barrel cortex) in vivo during the first postnatal week, we revealed that layer 4 (L4) cortical neurons within the same barrel fire synchronously in the absence of peripheral stimulation, creating a “patchwork” pattern of spontaneous activity corresponding to the barrel map. By generating transgenic mice expressing a genetically-coded calcium indicator GCaMP6s in thalamocortical axons, we showed that thalamocortical axons also demonstrated the spontaneous patchwork activity pattern. Patchwork activity was diminished by peripheral anesthesia but was mostly independent of self-generated whisker movements. The patchwork activity pattern largely disappeared during postnatal week 2, as even L4 neurons within the same barrel tended to fire asynchronously. This spontaneous L4 activity pattern has features suitable for circuit refinement in the neonatal barrel cortex.
This work was supported by JSPS KAKENHI grant numbers JP15K14322 and JP16H06143, the Takeda Science Foundation, and the Collaborative Research Project (2017-2923) of Brain Research Institute, Niigata University to H.M., and JSPS KAKENHI grant numbers JP16K14559, JP15H01454, and JP15H04263, and Grant-in Scientific Research on Innovation Areas “Dynamic Regulation of Brain Function by Scrap & Build System” (JP16H06459) from MEXT to T.I.
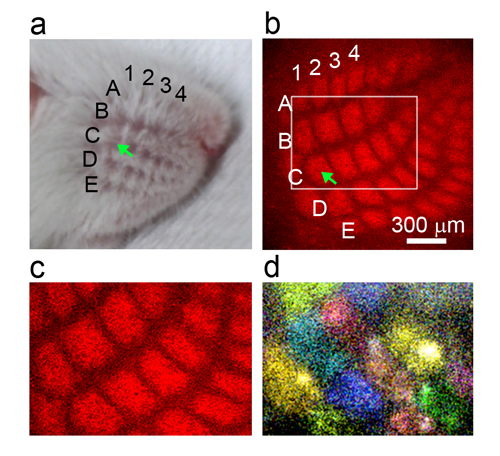
- Figure: Novel type of spontaneous activity found in the mouse somatosensory cortex layer 4 in neonates.
- (a) Five rows (A-E) of whiskers on the snout.
- (b) In the barrel cortex layer 4 (L4), barrels are arranged in a one-to-one correspondence with whiskers on the snout and process tactile information derived from individual whiskers. Each barrel contains hundreds of neurons processing sensory information from the corresponding whisker. For example, the C1 barrel (arrow) processes information from the C1 whisker (arrow in (a)). Barrels are labeled with a red fluorescent protein.
- (c) Higher magnification image of the white square region in (b).
- (d) Activity pattern of barrel cortex layer 4 at postnatal day 5, which looks like a “patchwork”. Each color represents a different activity event.















