Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
‘Eating with the eyes’ is hard-wired in the brain
Press release
Activation of the hypothalamic feeding centre upon visual prey detection
Akira Muto, Pradeep Lal, Deepak Ailani, Gembu Abe, Mari Itoh, Koichi Kawakami
Nature Communications 8, Article number: 15029 (2017) DOI:10.1038/ncomms15029
Pressrelease (In Japanese only)
Have you ever wondered why just seeing food can make your mouth start to water? By visualizing neuronal activity in specific areas of the zebrafish brain, scientists at the National Institute of Genetics (NIG) in Japan have revealed a direct link between visual perception of food and feeding motivation. The study, published in the April 20, 2017 issue of Nature Communications, suggests that “eating with the eyes” is deeply rooted in evolution.
This study was supported by JSPS KAKENHI Grant Numbers JP25290009 and JP25650120, and also partly supported by JSPS KAKENHI Grant Numbers JP15H02370 and JP16H01651, and NBRP from Japan Agency for Medical Research and Development (AMED). This work was also supported in part by the Center for the Promotion of Integrated Sciences (CPIS) of SOKENDAI.
Click here for article
https://www.eurekalert.org/pub_releases/2017-04/rooi-wt041717.php
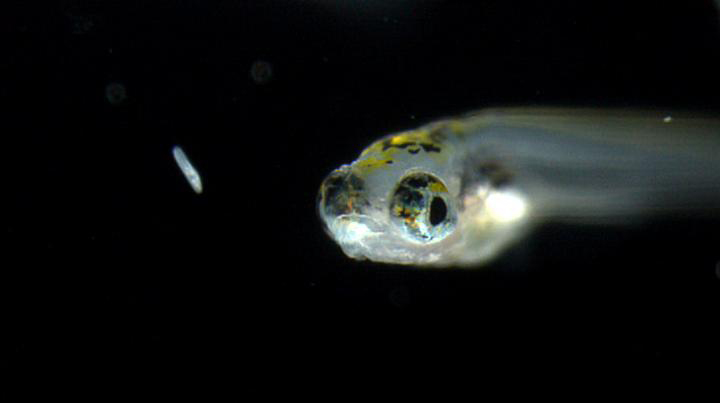
THIS IS A ZEBRAFISH LARVA TRYING TO CATCH PREY.
▶This study is based on the previous study.
Regulation of internode patterning and vein anastomosis in maize stems.
Experimental Farm / Nonomura Group
Mammalian Genetics Laboratory / Shiroishi Group
KNOTTED1 Cofactors, BLH12 and BLH14, Regulate Internode Patterning and Vein Anastomosis in Maize
Katsutoshi Tsuda, Maria-Jazmin Abraham-Juarez, Akiteru Maeno, Zhaobin Dong, Dale Aromdee, Robert Meeley, Toshihiko Shiroishi, Ken-ichi Nonomura and Sarah Hake.
The Plant Cell. published online April 5, 2017 DOI:10.1105/tpc.16.00967
Monocot stems lack the vascular cambium and instead have characteristic structures in which intercalary meristems generate internodes and veins remain separate and scattered. Developmental processes of these unique structures, however, have been poorly described. We found that maize BELL1-like homeodomain transcription factors, BLH12 and BLH14, have redundant but important roles in stem development. BLH12/14 interact with the shoot meristem regulator, KNOTTED1 (KN1) in vivo, and accumulate in overlapping domains in shoot meristems, young stems and provascular bundles. In addition to defects in the maintenance and development of various shoot meristems, blh12/14 double mutant showed unique abnormalities in the stem including shortened internodes and the reduced vein number. Detailed observation of BLH12/14 accumulation patterns and of stem inner structures using micro-computed tomography (CT) suggested that BLH12/14 (1) maintain intercalary meristems at the bottom of internodes to provide internodal cells as differentiated progenies, and (2) prevent precocious anastomosis of provascular bundles in young stems to ensure the production of sufficient independent veins. This work is the collaborative work between Katsutoshi Tsuda in Experimental Farm and Prof. Sarah Hake in University of California, Berkeley, and was supported by JSPS KAKENHI JP16K18637. Micro-CT scanning performed by Akiteru Maeno in Mammalian Genetics Lab enabled to capture the detailed view of inner stem structures and vein networks.
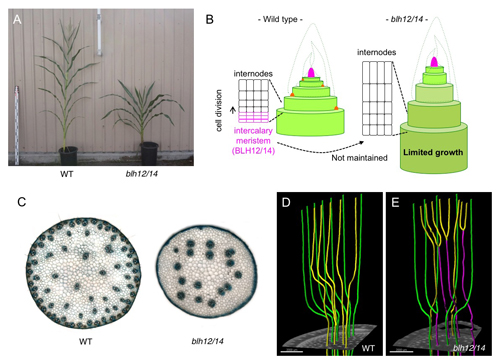
- (A) Wild-type (left) and blh12/14 (right) plants. blh12/14 double mutants show a dwarf phenotype.
- (B) Intercalary meristems are maintained by BLH12/14. In blh12/14, intercalary meristems were differentiated into internode and lost, leading to limited growth and dwarfness.
- (C) Transverse sections of wild-type (left) and blh12/14 double mutant (right) stems. blh12/14 had much fewer veins compared to wild type.
- (D) A vein network in the wild type stem extracted from micro CT data. Veins do not anastomose each other.
- (E) A vein network in the blh12/14 double mutant stem. Precociously anastomosed veins (magenta) were frequently observed.
Three new faculty have joined NIG as of April 1, 2017.
Three new faculty have joined NIG as of April 1, 2017.
professor
SAITO, Kuniaki : Invertebrate Genetics Laboratory
Associate Professor
KAWAMOTO, Shoko : Genetic Informatics Laboratory

- SAITO, Kuniaki
Professor

- KAWAMOTO, Shoko
Associate Professor
Assistant Professor












