Endoplasmic Reticulum-Mediated Microtubule Alignment Governs Cytoplasmic Streaming
Press release
Endoplasmic Reticulum-Mediated Microtubule Alignment Governs Cytoplasmic Streaming
Kenji Kimura, Alexandre Mamane, Tohru Sasaki, Kohta Sato, Jun Takagi, Ritsuya Niwayama, Lars Hufnagel, Yuta Shimamoto, Jean-François Joanny, Seiichi Uchida, and Akatsuki Kimura
Nature Cell Biology (2017) DOI:10.1038/ncb3490
Pressrelease (In Japanese only)
A research group lead by Drs. Kenji Kimura and Akatsuki Kimura at NIG revealed the self-organization mechanism of cytoplasmic streaming in Caenorhabditis elegans zygotes. Cytoplasmic streaming refers to a collective movement of cytoplasm observed in many cell types. The mechanism of meiotic cytoplasmic streaming (MeiCS) in C. elegans zygotes was puzzling as the direction of the flow is not predefined by cell polarity and occasionally reverses. The research group demonstrated that the endoplasmic reticulum (ER) network structure is required for the collective flow (Fig. 1). Using a combination of RNAi, microscopy, and image processing of C. elegans zygotes, the group devised a theoretical model, which reproduced and predicted the emergence and reversal of the flow. They proposed a positive feedback mechanism, where a local flow generated along a microtubule is transmitted to neighboring regions through the ER (Fig. 2). This, in turn, aligns microtubules over a broader area to self-organize the collective flow. The proposed model could be applicable to various cytoplasmic streaming phenomena in the absence of predefined polarity. The increased mobility of cortical granules by MeiCS correlated with the efficient exocytosis of the granules to protect the zygotes from osmotic and mechanical stresses.
The research was conducted as collaboration between NIG (Cell Architecture lab and Quantitative Mechanobiology lab), Kyushu University, Institut Curie (France) and EMBL (Germany).
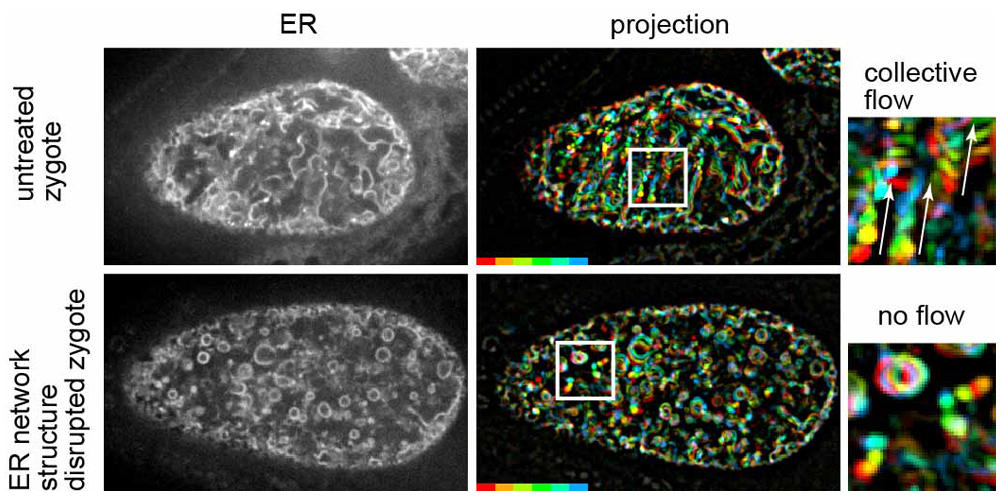
Fig. 1
Left panels show fluorescence confocal images of the ER during MeiCS in the C. elegans zygote (upper: untreated zygote, bottom: RNAi-treated zygote in which the network structure of the ER was fragmented). Middle panels show projections of sequential images of the ER. Fluorescence signals are colored to indicate their trajectories. Fragmentation of the ER network inhibited the flow. Boxed region is magnified on the right. White arrows indicate the flow direction.
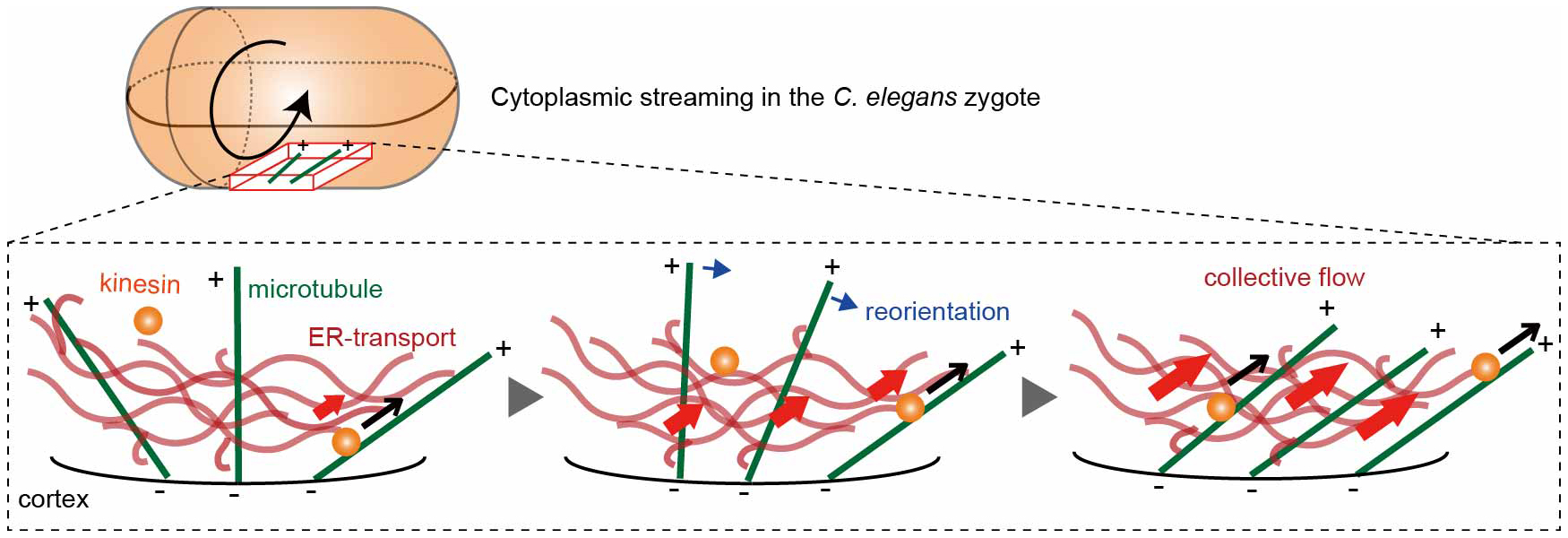
Fig. 2
ER-mediated positive feedback model. When kinesin transports the ER network along a cortical microtubule (black arrows), neighboring microtubules are biased to reorient (blue arrows) toward the ER motion, enhancing collective flow (red arrows). (−) and (+) signs indicate microtubule polarity.















