Wet-and-dry combination enables identification of functional enolase genes in silkworm
DBCLS
Comparative Genomics Laboratory
Advanced Genomics Center
Identification of functional enolase genes of the silkworm Bombyx mori from public databases with a combination of dry and wet bench processes
Akira Kikuchi, Takeru Nakazato, Katsuhiko Ito, Yosui Nojima, Takeshi Yokoyama, Kikuo Iwabuchi, Hidemasa Bono, Atsushi Toyoda, Asao Fujiyama, Ryoichi Sato and Hiroko Tabunoki
BMC Genomics, 18:83, 2017 DOI:10.1186/s12864-016-3455-y
Researchers can now easily obtain genomic information from online databases. However, many incorrectly annotated genes are included in these databases, which can prevent the correct interpretation of subsequent functional analyses. To address this problem, we used a combination of dry and wet bench processes to identify functional enolases in the silkworm Bombyx mori.
To identify enolase sequences in B. mori, we performed a HMM search of public databases. We found five enolase sequences, which we then characterized using RNA-seq analysis, cDNA cloning, and RT-PCR. Finally, we determined that three enolase genes in B. mori were functional. Our strategy could be helpful for the detection of minor genes and functional genes in non-model organisms from public databases.
Prof. Atsushi Toyoda and Prof. Asao Fujiyama (Center for Information Biology) contributed to sequencing testis transcriptome of B. mori. Dr. Takeru Nakazato and Dr. Hidemasa Bono (Database Center for Life Science) contributed to this work in dry analysis.
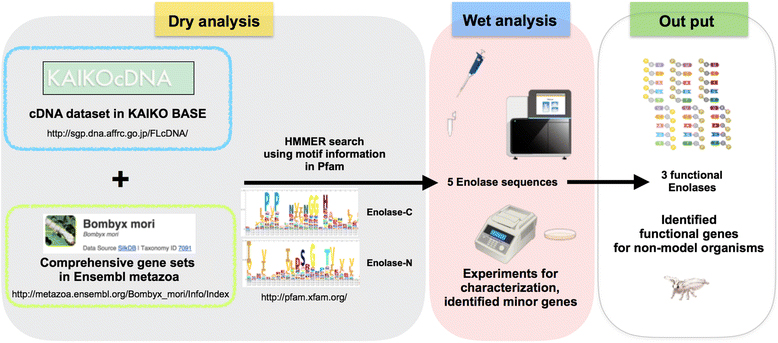
Combining dry and wet bench processes to identify functional enolases in the silkworm B. mori. To identify enolase sequences in B. mori, we performed a HMM search of public databases. We found five enolase sequences, which we then characterized using RNA-seq analysis, cDNA cloning, and RT-PCR. Finally, we determined that three enolase genes in B. mori were functional.















