Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Specific DNA-binding activity of bacterial condensin
Microbial Genetics Laboratory / Niki Group
In vitro topological loading of bacterial condensin MukB on DNA, preferentially single-stranded DNA rather than double-stranded DNA
Hironori Niki, and Koichi Yano
Scientific Reports 6, Article number: 29469 (2016) DOI:10.1038/srep29469
Condensin is the major driving force in the segregation of daughter chromosomes in prokaryotes. Core subunits of condensin belong to the SMC protein family, whose members are characterized by a unique ATPase activity and dimers with a V-shaped structure. The V-shaped dimers might close between head domains, forming a ring structure that can encircle DNA. Indeed, cohesin, which is a subfamily of SMC proteins, encircles double-stranded DNA to hold sister chromatids in eukaryotes. However, the question of whether or not condensin encircles the chromosomal DNA remains highly controversial. Here we report that MukB binds topologically to DNA in vitro, and this binding is preferentially single-stranded DNA (ssDNA) rather than double-stranded DNA. The binding of MukB to ssDNA does not require ATP. In fact, thermal energy enhances the binding. The non-SMC subunits MukF and MukE did stimulate the topological binding of MukB, although they hindered DNA-binding of MukB. Recent reports on the distribution of condensin in genomes reveal that actively transcribed genes in yeast and humans are enriched in condensin. In consideration of all these results, we propose that the binding specificity of condensin to chromosome is provided not by the DNA sequence but by the DNA structure, which is ssDNA.
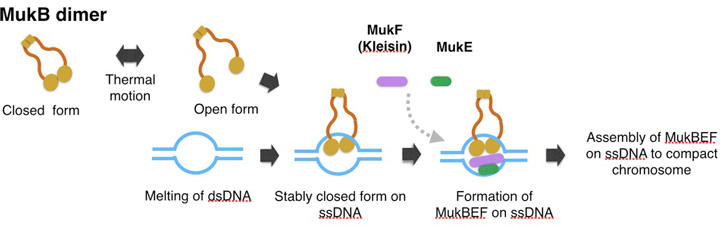
A model of topological binding of MukB in E. coli cells.
We hypothesize that the arms of MukB dimers are flexible and change between the open form or closed form depending on the thermal fluctuation. Occasionally the open form captures DNA, and then changes into the closed form. After the MukB dimer captures ssDNA, the closed form would become static because a part of the inner interface of a MukB globular domain interacts with ssDNA. Thus MukB would keep the captured DNA steady inside the ring of the dimer. Further, the MukB-DNA complex might be strengthened by MukEF, and then each of the MukBEF-DNA complexes would be assembled to compact chromosomal DNA. We infer that ATP hydrolysis is required for dissociation of the MukBEF-DNA complex from the assembled complexes.
A simple telomere labeling method in tissue sections.
Press release
Telomere Visualization in Tissue Sections using Pyrrole–Imidazole Polyamide Probes
Asuka Sasaki, Satoru Ide, Yusuke Kawamoto, Toshikazu Bando, Yukinori Murata, Mari Shimura, Kazuhiko Yamada, Akiyoshi Hirata, Kiyoshi Nokihara, Tatsumi Hirata, Hiroshi Sugiyama, Kazuhiro Maeshima
Scientific Reports 6: 29261 (2016) DOI:10.1038/srep29261
Press Release (Only in Japanese)
Pyrrole–Imidazole (PI) polyamides bind to specific DNA sequences in the minor groove with high affinity. Specific DNA labeling by PI polyamides does not require DNA denaturation with harsh treatments of heat and formamide and has the advantages of rapid and less disruptive processing. Previously, we developed tandem hairpin PI polyamide probes (TH59 series), which label telomeres in cultured cell lines more efficiently than conventional methods, such as fluorescence in situ hybridization (FISH). Here, we demonstrate that a TH59 derivative, HPTH59-b, along with immunostaining for specifying cell types in the tissues, visualizes telomeres in mouse and human tissue sections. Quantitative measurements of telomere length with single-cell resolution suggested shorter telomeres in the proliferating cell fractions of tumor than in non-tumor tissues. Thus, PI polyamides are a promising alternative for telomere labeling in clinical research, as well as in cell biology.
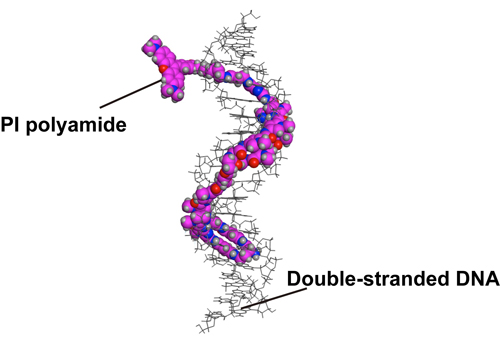
Figure1. A structural model of HPTH59-b binding to DNA.
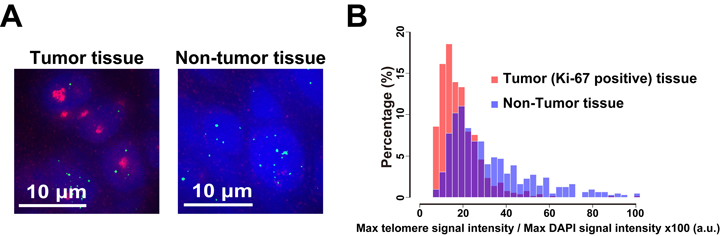
Figure2. Different telomere lengths between human tumor and non-tumor tissue sections. (A) Frozen sections of esophageal tumor/non-tumor tissue stained with DAPI (blue), anti-Ki-67 (growth marker; green) antibody, and HPTH59-b (red). Distribution histograms of telomere signal intensities in tumor and non-tumor tissue sections.
New assistant professor joins NIG
New assistant professor joins NIG as of July 1, 2016.
Daisuke TAKAO: Division of Centrosome Biology, Kitagawa Group















