Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Turtle’s and chicken’s brains are similar!?
Division of Brain Function • Hirata Group
A common developmental plan for neocortical gene-expressing neurons in the pallium of the domestic chickenGallus gallus domesticus and the Chinese softshell turtle Pelodiscus sinensis.
I. K. Suzuki and T. Hirata Front. Neuroanat.(2014) 8 20 doi: 10.3389/fnana.2014.00020The highly evolved neocortex is a marked characteristic of mammals. It develops as layers, in which neurons are sequentially added from deep to the surface in a birth-order-dependent manner. Evolutionary processes of the layered neocortex have been a long mystery. We recently reported that the chicken brain contains neuron subtypes that are similar to deep and upper layer neurons in the mammalian neocortex . However, the comparison of only two distant animal groups was not sufficient to specify potential evolutional changes, and similar analyses of other animal groups have been awaited. In this study, we performed gene expression analyses in the softshell turtle brain, which is often referred to as an ancestral form of amniote brains. The turtle brain contains a single neuronal layer, exhibiting a clear difference from the chicken brain. Our expression analyses of orthologs of neocortical layer marker genes highlighted similarities in neuronal arrangements between the two species. Specifically, in both of the species, deep layer neurons are positioned in the medial part, whereas upper layer neurons are positioned in the lateral part. Thus, this medio-lateral neuronal arrangement appears to be an ancestral mode in sauropsids. The results support our hypothesis of an ancient origin of neocortical neuron subtypes. We assume that the generality of birth-order-dependent mechanisms for specification of neuron subtypes probably underlies the their unexpected conservation among diverse amniote species.
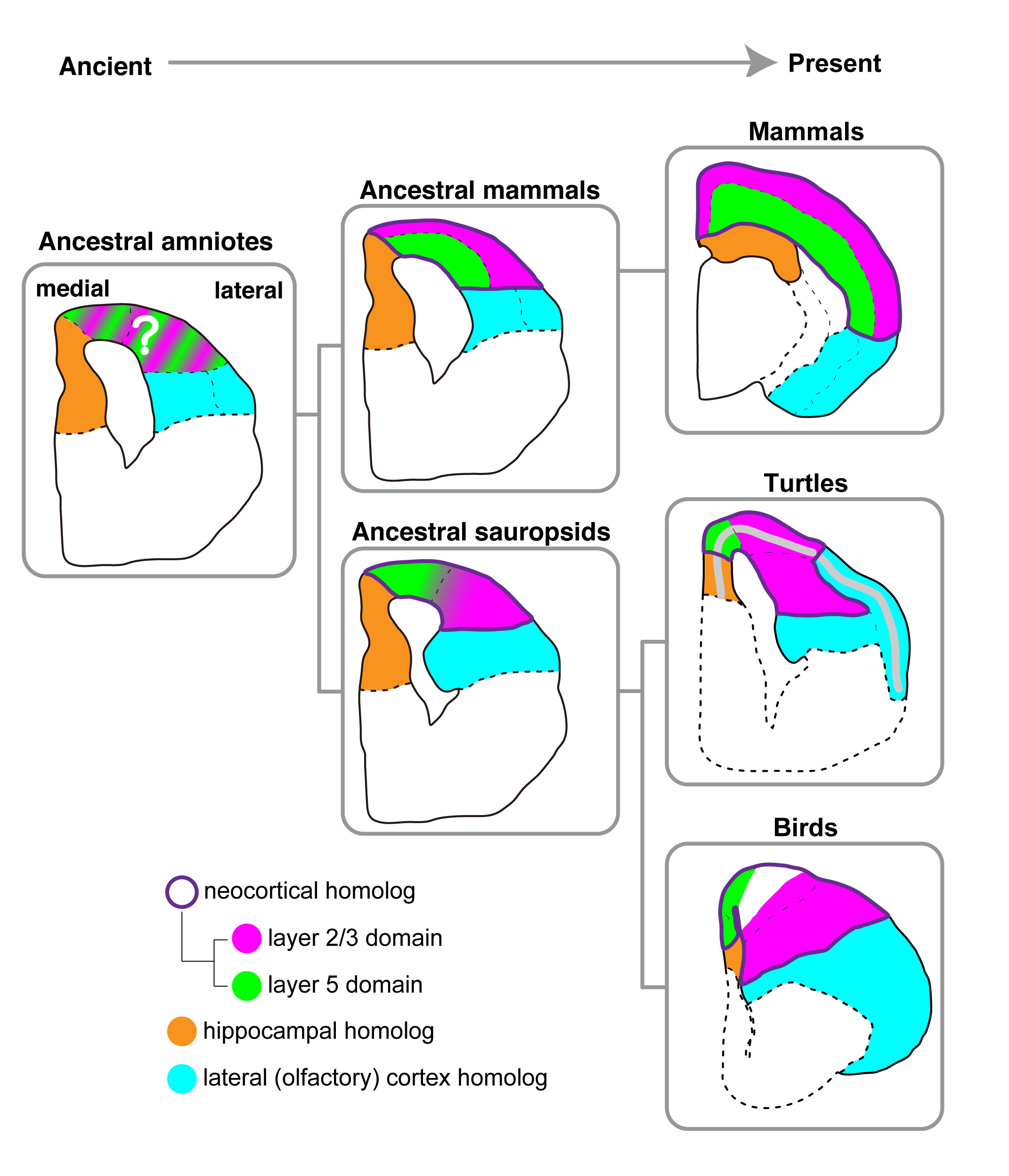
Evolutionary model of brains in amniotes
Development of a freeware that enables automatic characterization of social behavior in mice
Press release
A male-specific QTL for social interaction behavior in mice mapped with automated pattern detection by a hidden Markov model incorporated into newly developed freeware
Toshiya Arakawa1, Akira Tanave1, Shiho Ikeuchi, Aki Takahashi, Satoshi Kakihara, Shingo Kimura, Hiroki Sugimoto, Nobuhiko Asada, Toshihiko Shiroishi, Kazuya Tomihara, Takashi Tsuchiya, Tsuyoshi Koide. ※1 equally contributed.
Journal of Neuroscience Methods Available online 21 April 2014 doi:10.1016/j.jneumeth.2014.04.012
Notwithstanding the importance of effective approaches to analyze social interaction between animals in experimental settings, the methods that are currently available to do this rely predominantly on human observation. This makes large-scale studies of social interaction behavior difficult.
In this paper, we report the development of freeware, called DuoMouse, that enables video recording to track the movements of two mice from a movie file, analysis of behavioral states using a hidden Markov model (HMM), and visualization of the results. We used this software to compare social behavior in consomic and subconsomic strains, and to map a genetic locus responsible for differences in social interaction between the strains. We report a locus that increases social interaction only in males and that lies within a 24.6-Mb region of chromosome 6.
We propose that the method applied in the present study is very efficient and useful for large-scale genetic and pharmacological studies of social behavior in mice. All of the software is open-source, and the source code will also be provided for further development by others.
(https://zenodo.org/records/12577797)
This research was supported by the Research Organization of Information and Systems, Transdisciplinary Research Integration Center, JSPS KAKENHI Grant Numbers 23650243 and 25116527, and NIG Cooperative Research Program (2010-A40, 2012-A85).
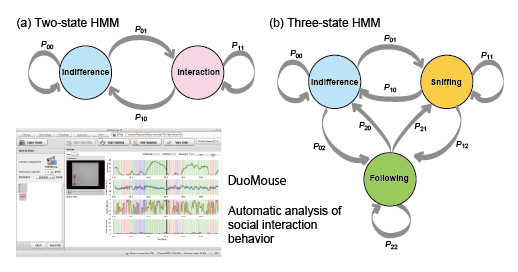
Diagram showing the Markov transition probabilities for the two-state HMM and three-state HMM. The image showing one of the pages of the freeware, DuoMouse.
Development of the lateral line canal system through a bone remodeling process in zebrafish
Division of Molecular and Developmental Biology • Kawakami Group
Development of the lateral line canal system through a bone remodeling process in zebrafish.
Hironori Wada, Miki Iwasaki, Koichi Kawakami Dev. Biol., in press, doi: 10.1016/j.ydbio.2014.05.004The lateral line system of teleost fish is composed of mechanosensory receptors (neuromasts), some of which are superficial whereas others are embedded in canals running under the skin. Canal diameter and size of canal neuromasts are correlated with increasing body size, thus providing a very simple system to investigate the mechanism underlying the coordination between organ growth and body size. Here, we examine the development of the trunk lateral line canal system in zebrafish. We demonstrate that trunk canals originate from scales through a bone remodeling process. We suggest that the bone remodeling process is essential for normal growth of canals and of canal neuromasts. Moreover, we show that the presence of lateral line cells is required for the formation of canals, suggesting the existence of mutual interactions between the sensory system and surrounding connective tissues.
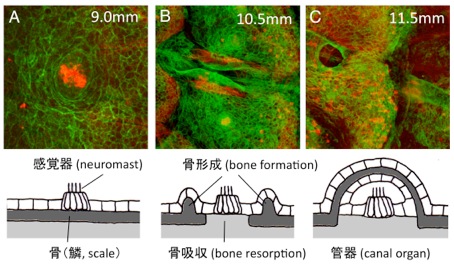
(A) The lateral line sense organ, the neuromast, is located on the skin surface in embryonic stages. (B) When fish undergo metamorphosis, a pair of ridges extends outward from the scale. At the same time, bony elements under the neuromast gradually disappear. (C) The neuromast becomes embedded in the canal running under the scale in adult fish.
Mudi•‘one-click’ identification of causative on web.
Genome Informatics Laboratory • Nakamura Group Division of Cytogenetics • Kobayashi Group
Mudi, a web tool for identifying mutations by bioinformatics analysis of whole-genome sequence.
Naoko Iida, Fumiaki Yamao, Yasukazu Nakamura, and Tetsushi Iida Genes to Cells 28 APR 2014 DOI:10.1111/gtc.12151Genetics is a powerful approach to discovery of the molecular mechanisms underlying biological phenomena. However, experiments for identification of mutations by classical genetics is a time-consuming and laborious process. Modern whole-genome sequencing, coupled with bioinformatics analysis, has enabled fast and cost-effective mutation identification. However, for many experimental researchers, bioinformatics analysis is still a difficult aspect of whole-genome sequencing. To address this issue, we developed a browser-accessible and easy-to-use bioinformatics tool called Mutation discovery (Mudi; http://naoii.nig.ac.jp/mudi_top.html). Users can run Mudi analysis with ‘one-click’ operation in a web browser after uploading genome resequencing data. Mudi simplifies analysis of whole-genome sequencing and thereby expands the possibilities for systematic forward-genetic approaches in various organisms.
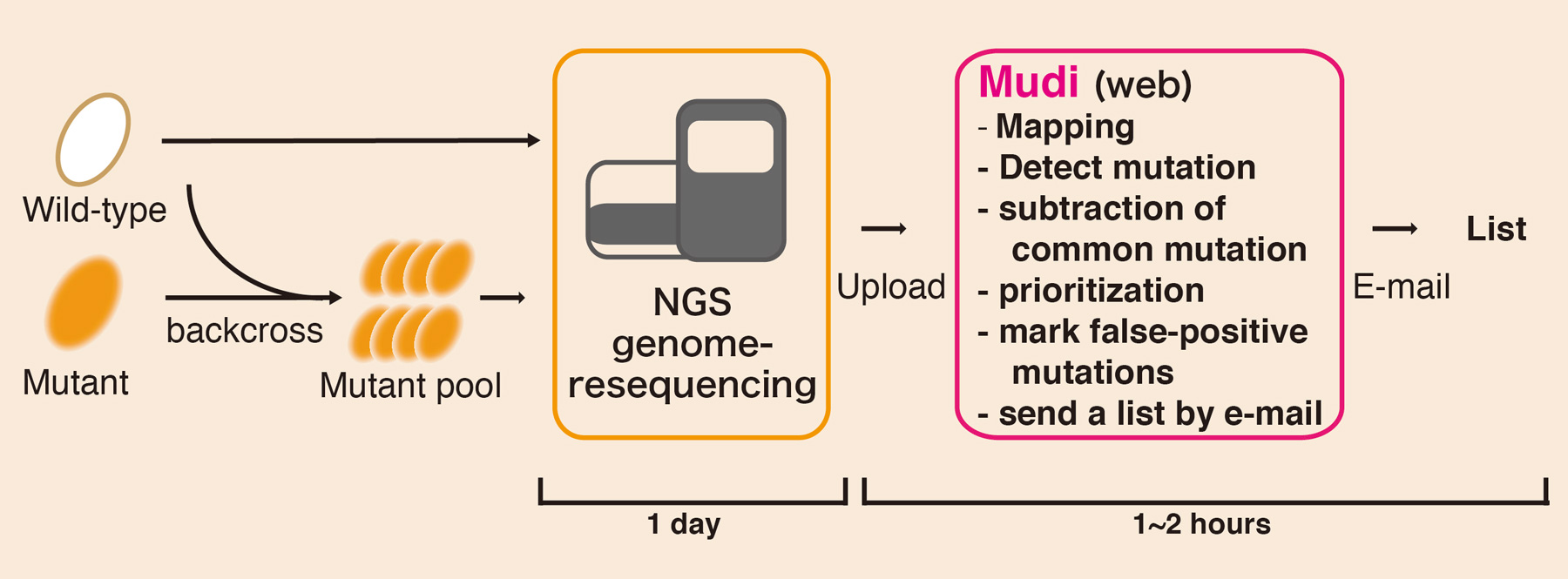
Workflow of “Mudi” system. Genomic DNA is prepared from pooled mutants by pooled-linkage analysis. Mudi calls prioritized mutation candidates by bioinformatics analysis using genome-resequencing data.
A cluster of four QTLs for behavior segregated by new method using a statistical regression model
Mouse Genomics Resource Laboratory (MGRL) • Koide Group
Segregation of a QTL cluster for home-cage activity using a new mapping method based on regression analysis of congenic mouse strains
Shogo Kato, Ayako Ishii, Akinori Nishi, Satoshi Kuriki, Tsuyoshi Koide Heredity. advance online publication 30 April 2014; doi:10.1038/hdy.2014.42Genetic mapping using congenic strains is one of the most powerful methods available to show the existence of QTLs in the genomic regions substituted into recipient strains. In this case, genetic mapping can be conducted by making a series of congenic strains that cover genomic regions that partially overlap with the adjacent congenic strains. In the analysis of the phenotype using a series of congenic strains, however, the phenotype shows a variety of levels, which indicates the existence of multiple QTLs in the mapped chromosomal regions. These results show that the precise genetic mapping of fine QTLs is extremely difficult using current methods.
Dr. Tsuyoshi Koide’s group collaborated with Drs. Shogo Kato and Satoshi Kuriki in the Institute of Statistical Mathematics and established a new method for mapping multiple QTLs using data from a series of congenic strains by applying a regression model for the analysis of total home-cage activity in mice. The results of the analysis identified four significant QTLs in a 14.5 Mb genomic region. Among these, three have negative effects but one has a positive effect on total home-cage activity. In further analysis of recombinants obtained from the congenic strains, we confirmed the existence of the QTL that has a positive effect on the activity as well as another QTL that suppresses the effect of this positive QTL. These results clarify for the first time the association of a complex genetic mechanism in the QTL cluster on chromosome 6 with the regulation of home-cage activity.
This work was supported by the Research Organization of Information and Systems, Transdisciplinary Research Integration Center, JSPS KAKENHI (Grant Numbers 23650243 and 25116527), and Yamada Science Foundation.
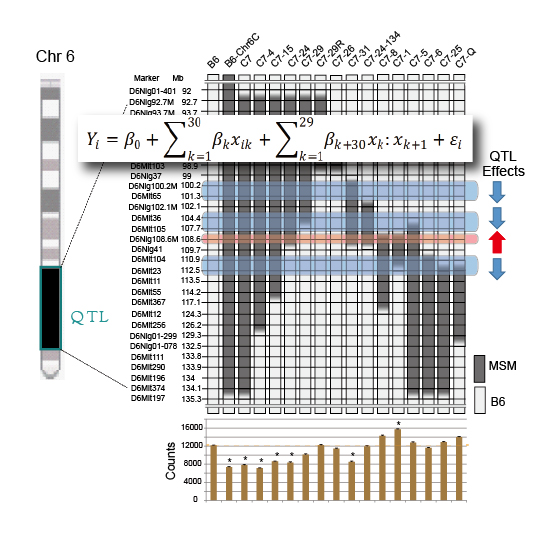
A cluster of four QTLs for behavior segregated by new method using a statistical regression model. The data of genomic regions carried in the congenic strains as well as the data of home-cage activity exhibited by congenic strains are analyzed with regression model. The study revealed four QTLs which are clustered in a small genomic region.















